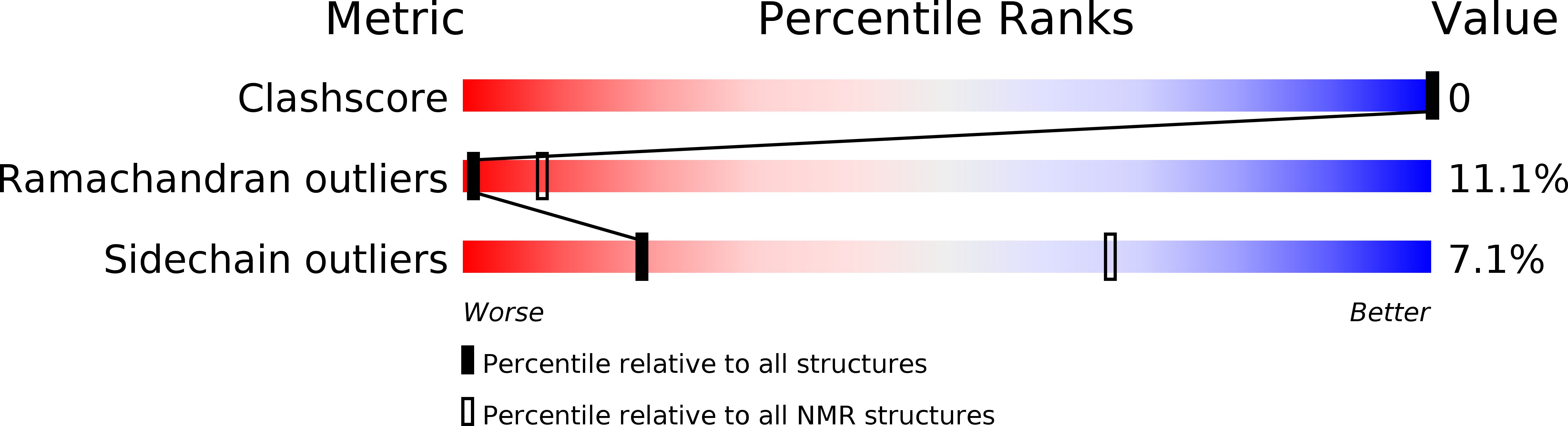
Deposition Date
2012-01-06
Release Date
2012-08-08
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2LNY
Keywords:
Title:
ShB peptide structure bound to negatively charged lipid-bilayer after Molecular Dynamics refinement
Biological Source:
Source Organism(s):
artificial gene (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
1
Conformers Submitted:
1
Selection Criteria:
target function