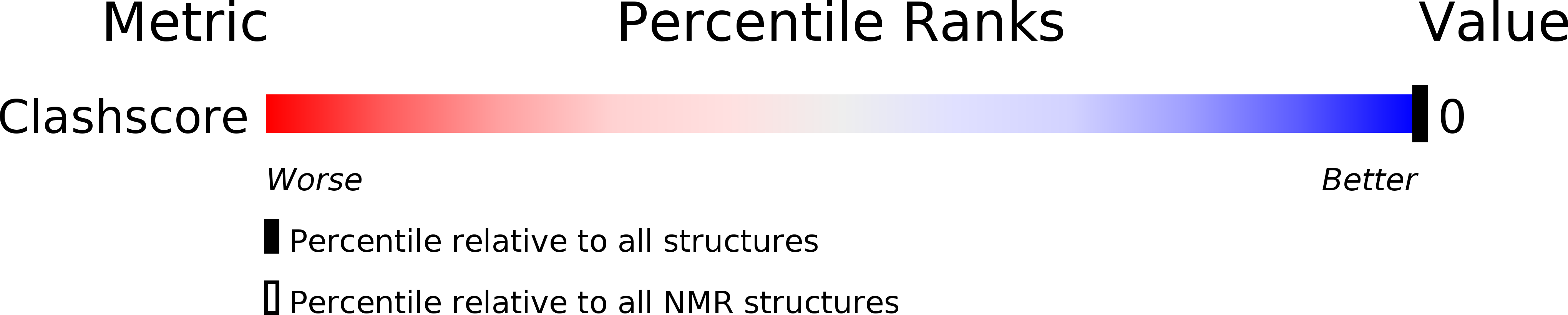
Deposition Date
2011-07-18
Release Date
2012-06-27
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2LFX
Keywords:
Title:
Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dT
Method Details:
Experimental Method:
Conformers Calculated:
10
Conformers Submitted:
1
Selection Criteria:
back calculated data agree with experimental NOESY spectrum