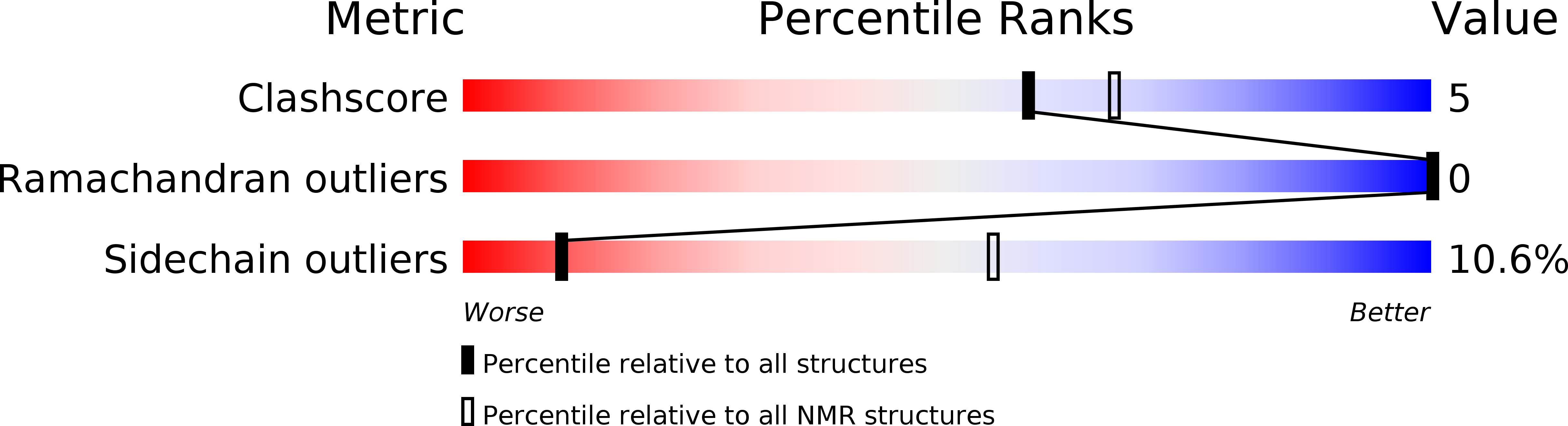
Deposition Date
2011-06-28
Release Date
2011-07-13
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2LF3
Keywords:
Title:
Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A
Biological Source:
Source Organism(s):
Pseudomonas syringae pv. maculicola (Taxon ID: 59511)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy