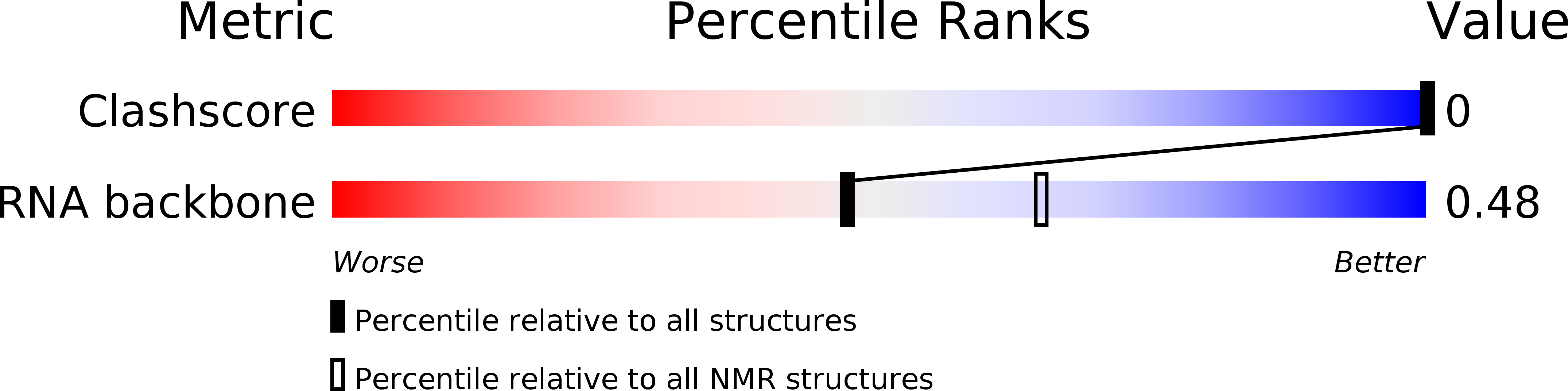
Deposition Date
2011-01-07
Release Date
2011-02-23
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2L8C
Keywords:
Title:
NMR Spectroscopy and Molecular Dynamics Simulation of r(CCGCUGCGG)2 Reveal a Dynamic UU Internal Loop Found in Myotonic Dystrophy Type 1 - UU pair with zero hydrogen bond pairs
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
Conformers Calculated:
25
Conformers Submitted:
1
Selection Criteria:
structures with acceptable covalent geometry