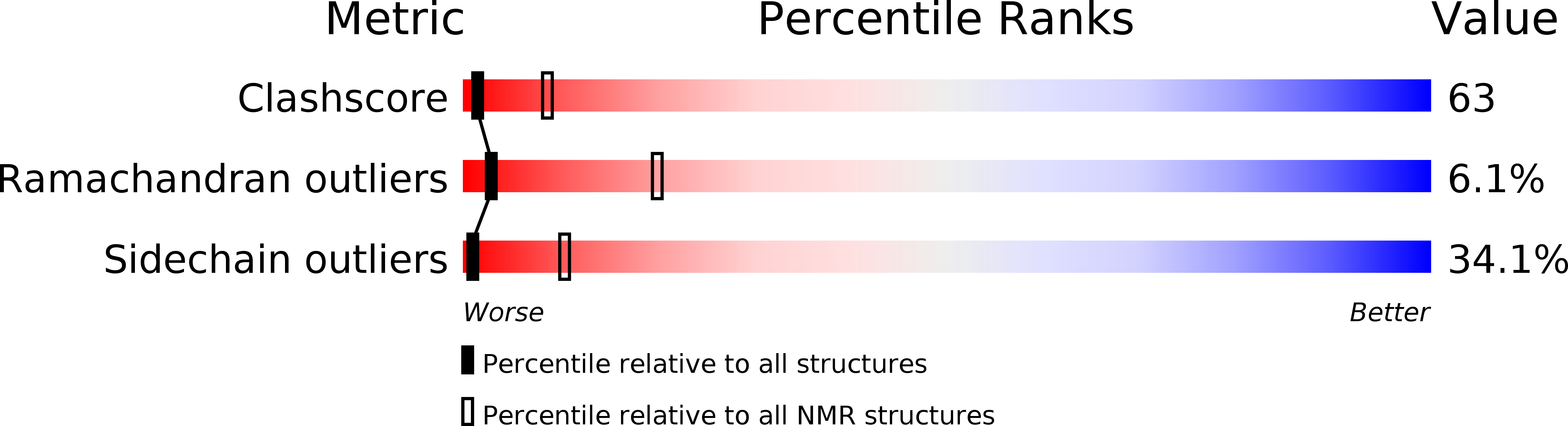Deposition Date
2010-11-17
Release Date
2011-03-23
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2L6B
Keywords:
Title:
NRC consensus ankyrin repeat protein solution structure
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
80
Conformers Submitted:
20
Selection Criteria:
back calculated data agree with experimental NOESY spectrum