Deposition Date
2010-07-28
Release Date
2010-10-27
Last Version Date
2024-05-01
Entry Detail
PDB ID:
2L1F
Keywords:
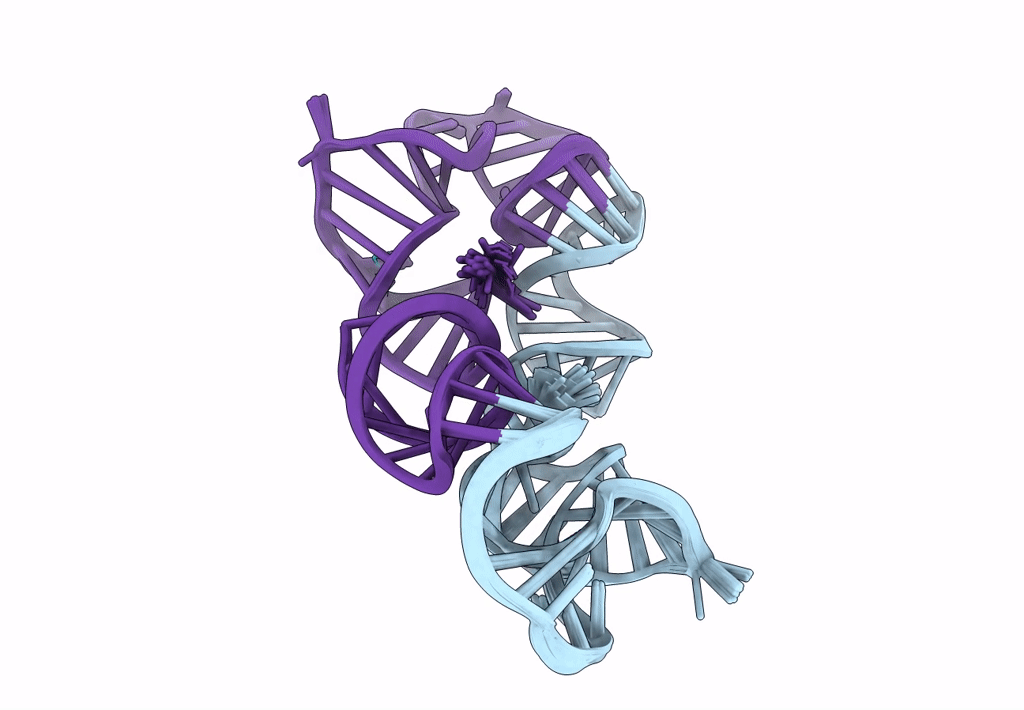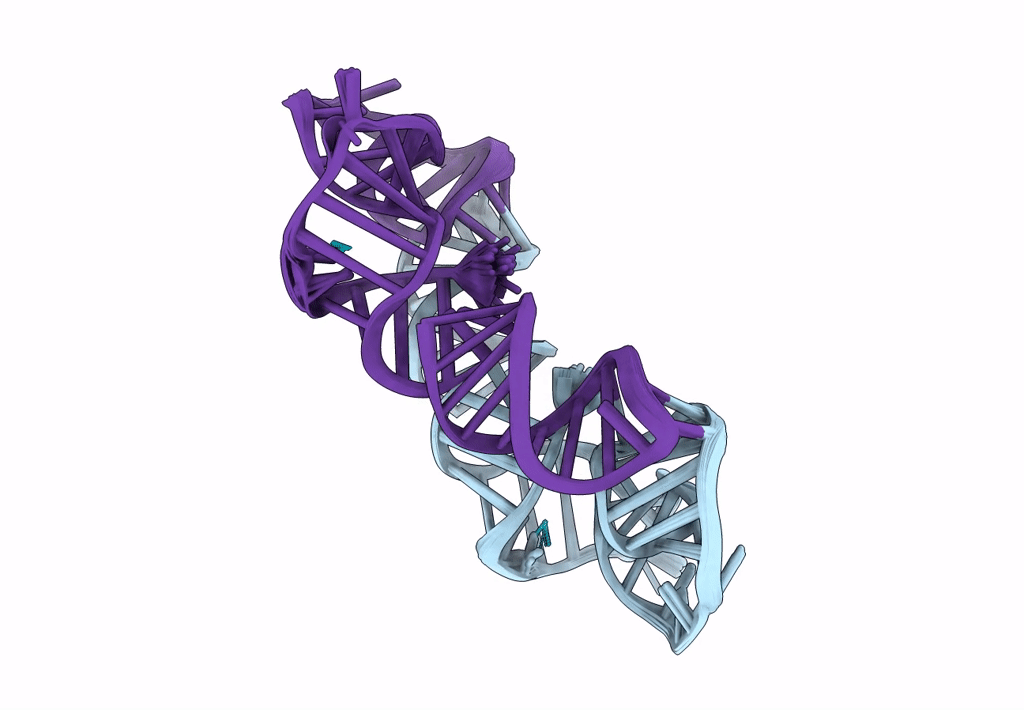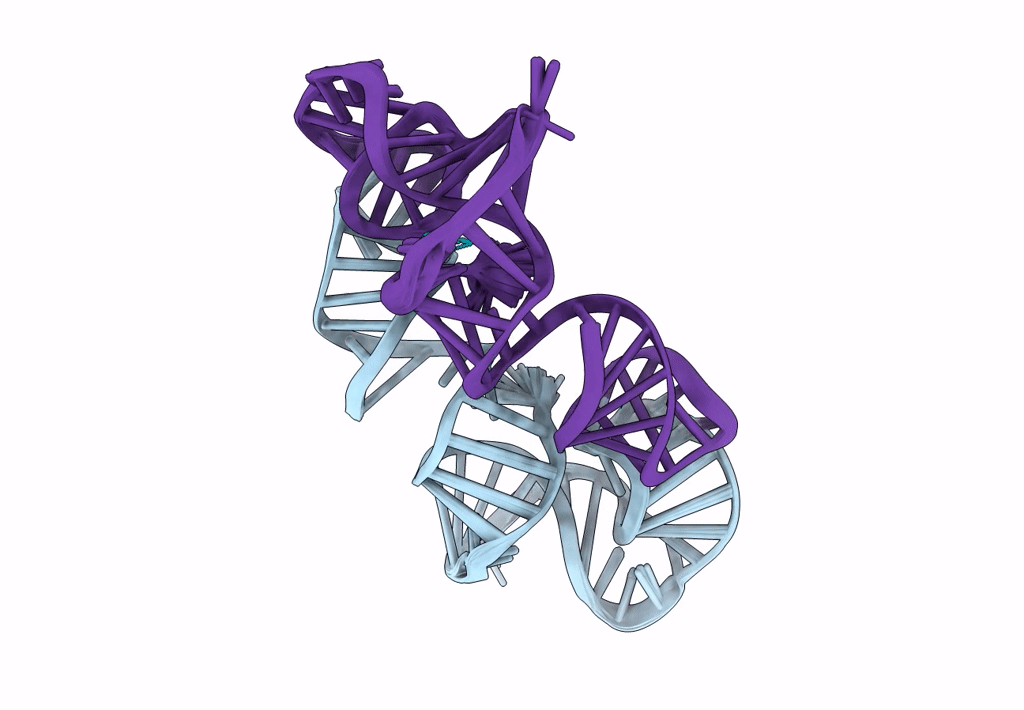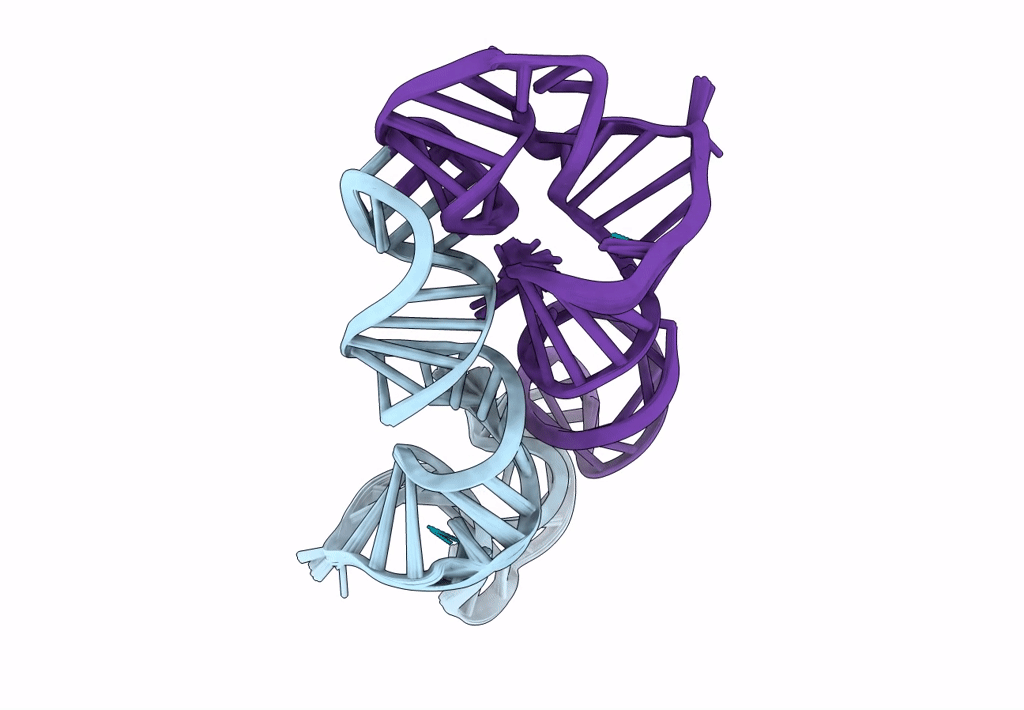
Title:
Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography
Biological Source:
Source Organism(s):
Moloney Murine Leukemia Virus (Taxon ID: 11801)
Method Details:
Experimental Method:
Conformers Calculated:
340
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy


