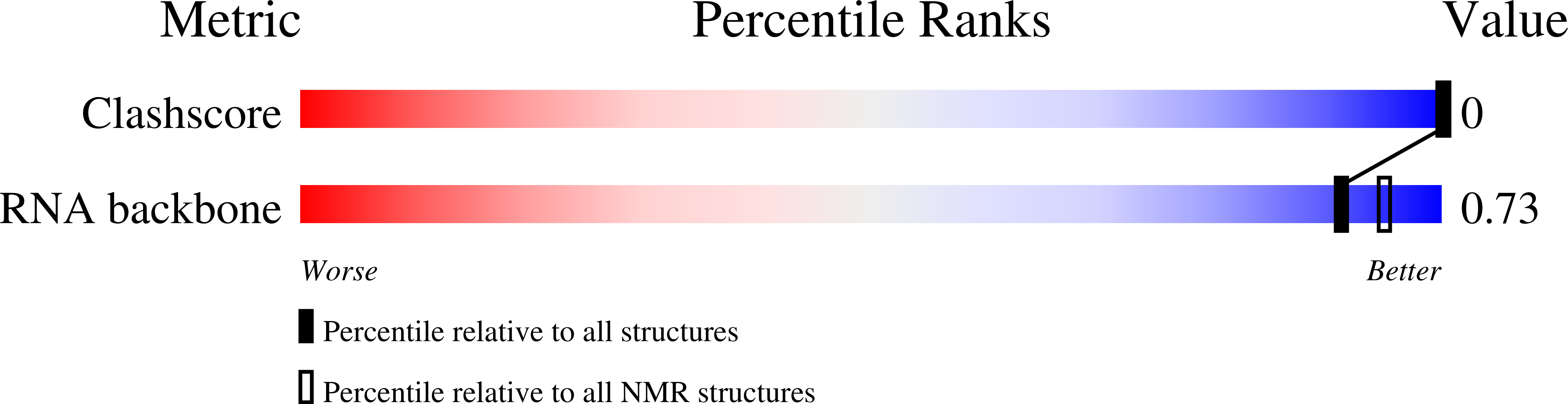
Deposition Date
2010-05-13
Release Date
2010-06-02
Last Version Date
2024-05-22
Entry Detail
PDB ID:
2KY1
Keywords:
Title:
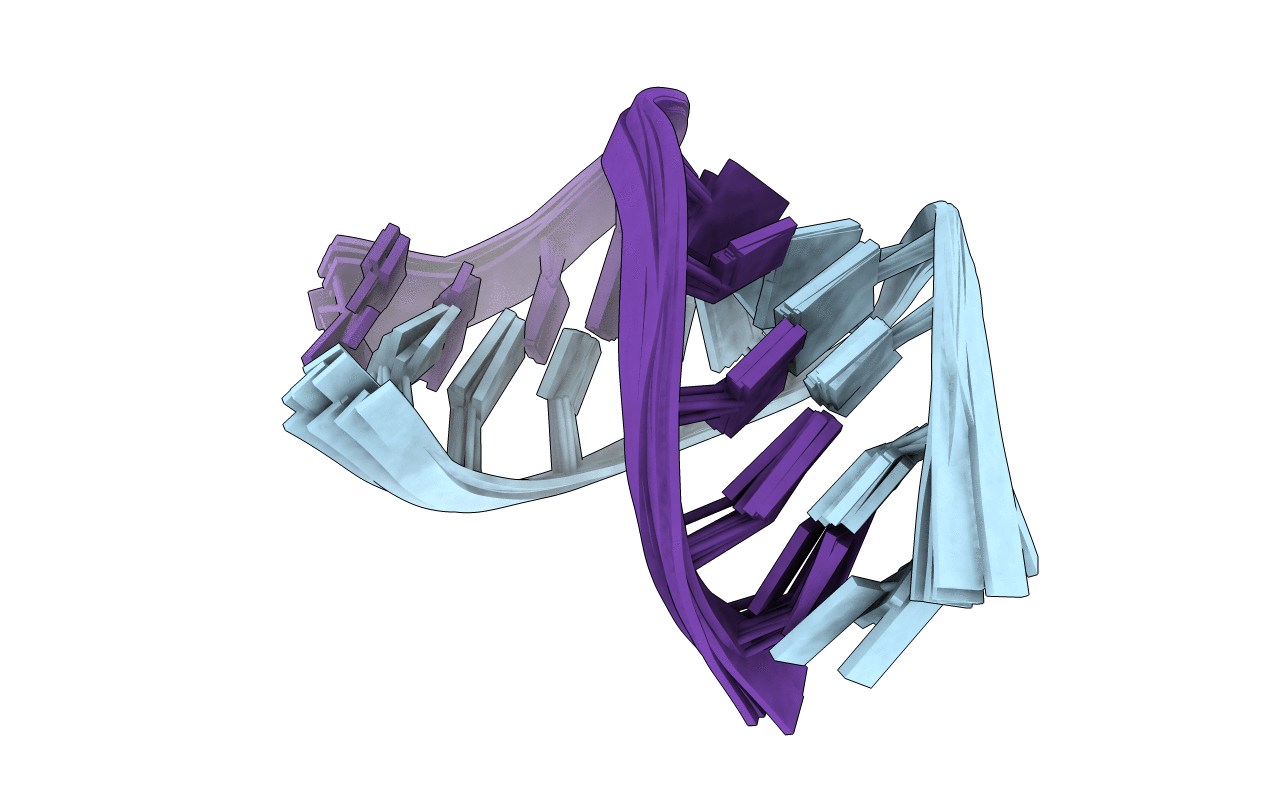
The Structure of RNA Internal Loops with Tandem AG Pairs: 5'UAGA/3'AGAU
Method Details:
Experimental Method:
Conformers Calculated:
40
Conformers Submitted:
10
Selection Criteria:
structures with the least restraint violations