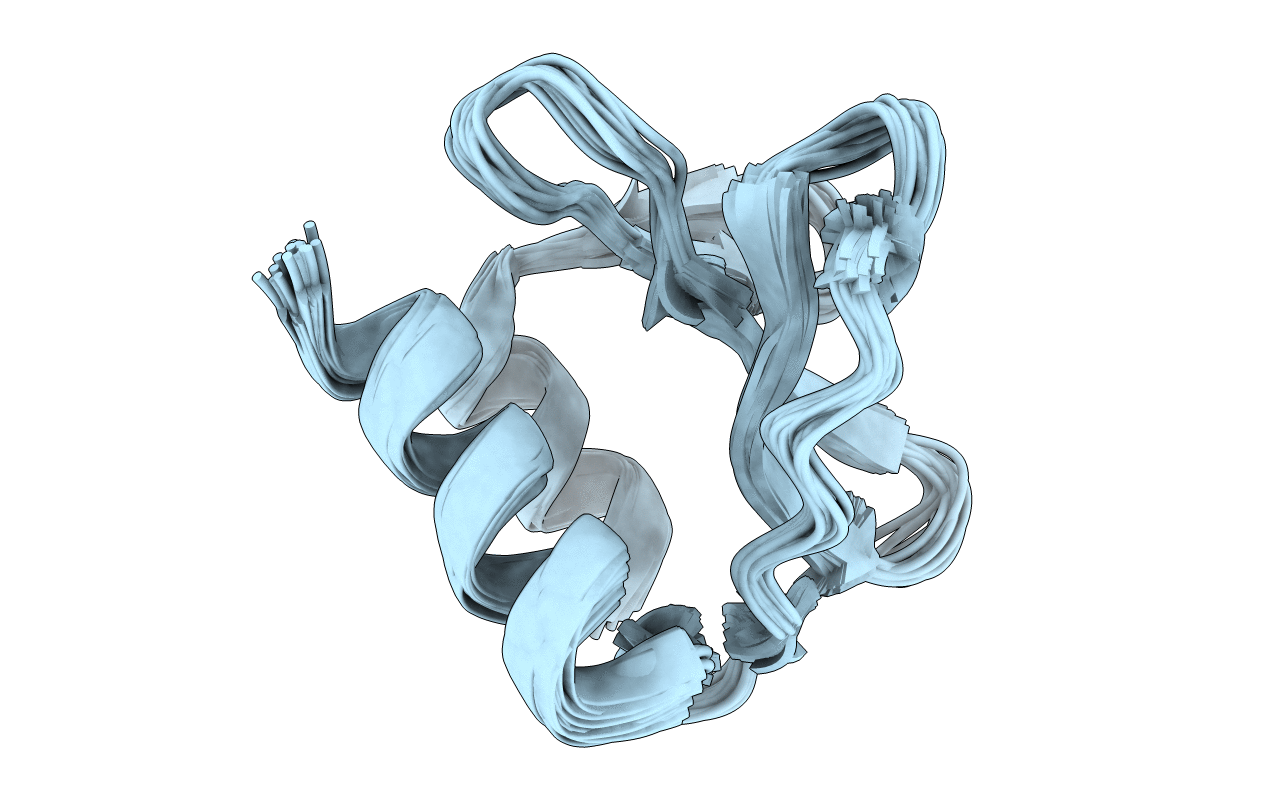
Deposition Date
2009-11-19
Release Date
2010-02-02
Last Version Date
2024-05-01
Entry Detail
PDB ID:
2KQW
Keywords:
Title:
SARS coronavirus-unique domain (SUD): Three-domain molecular architecture in solution and RNA binding. II: Structure of the SUD-C domain of SUD-MC
Biological Source:
Source Organism(s):
SARS coronavirus (Taxon ID: 227859)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
80
Conformers Submitted:
20
Selection Criteria:
target function