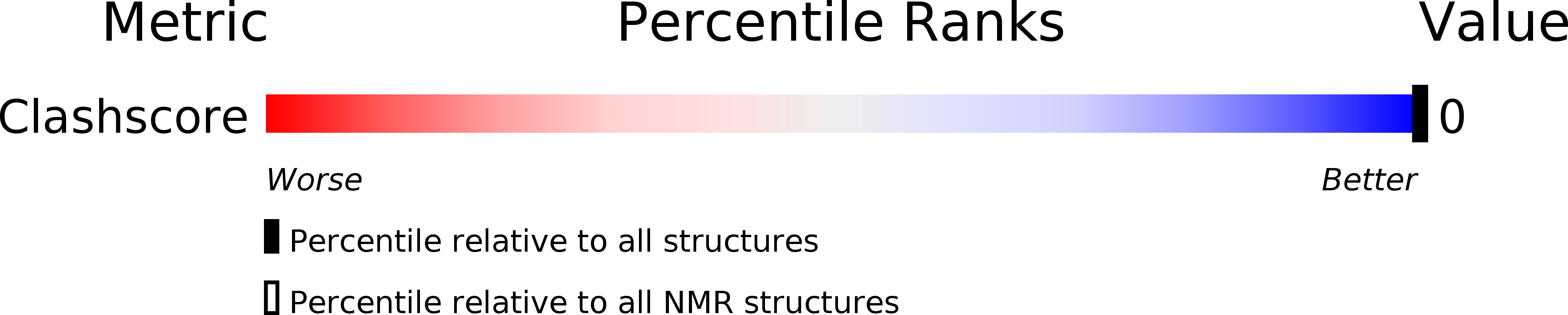
Deposition Date
2009-11-12
Release Date
2009-12-01
Last Version Date
2024-05-01
Entry Detail
PDB ID:
2KQO
Keywords:
Title:
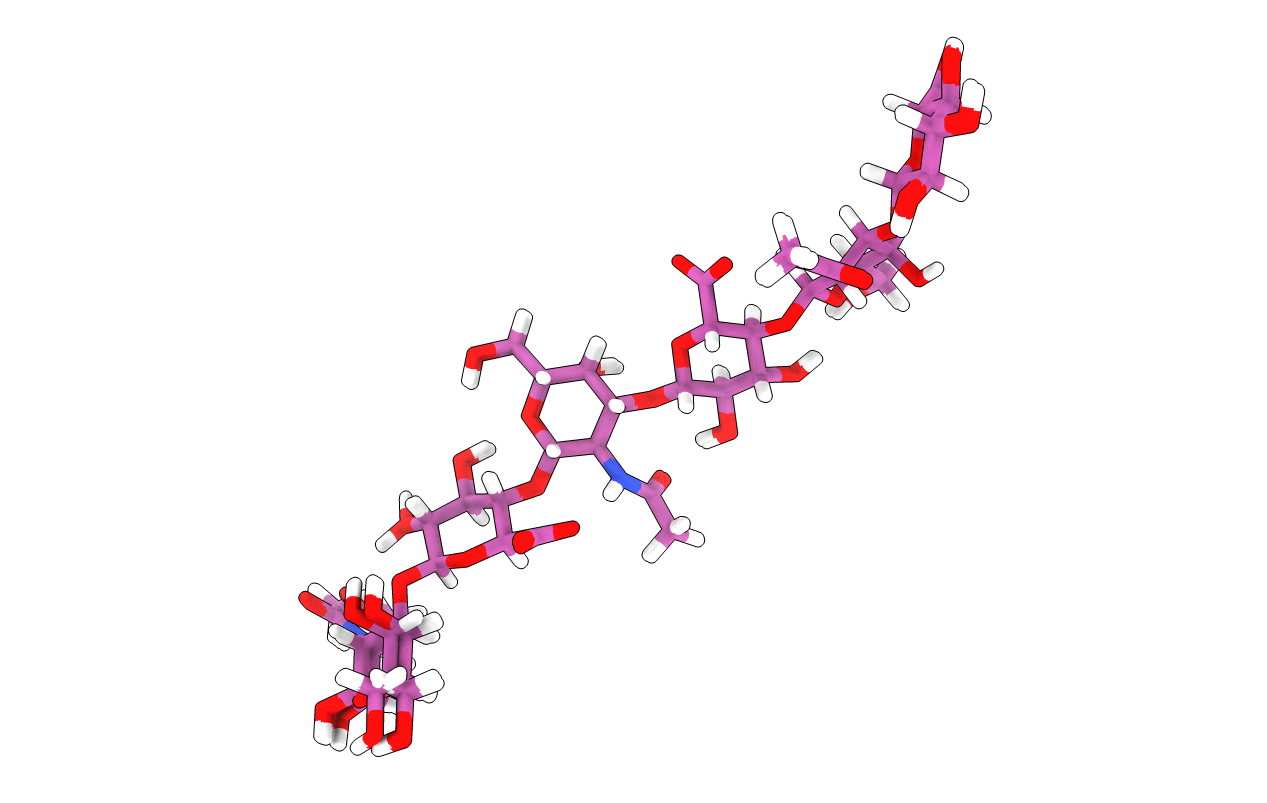
A 3D-structural model of unsulphated chondroitin from high-field NMR: 4-sulphation has little effect on backbone conformation
Method Details:
Experimental Method:
Conformers Calculated:
250
Conformers Submitted:
25
Selection Criteria:
structures with the lowest energy