Deposition Date
2009-01-30
Release Date
2009-05-19
Last Version Date
2024-11-20
Entry Detail
PDB ID:
2KEI
Keywords:
Title:
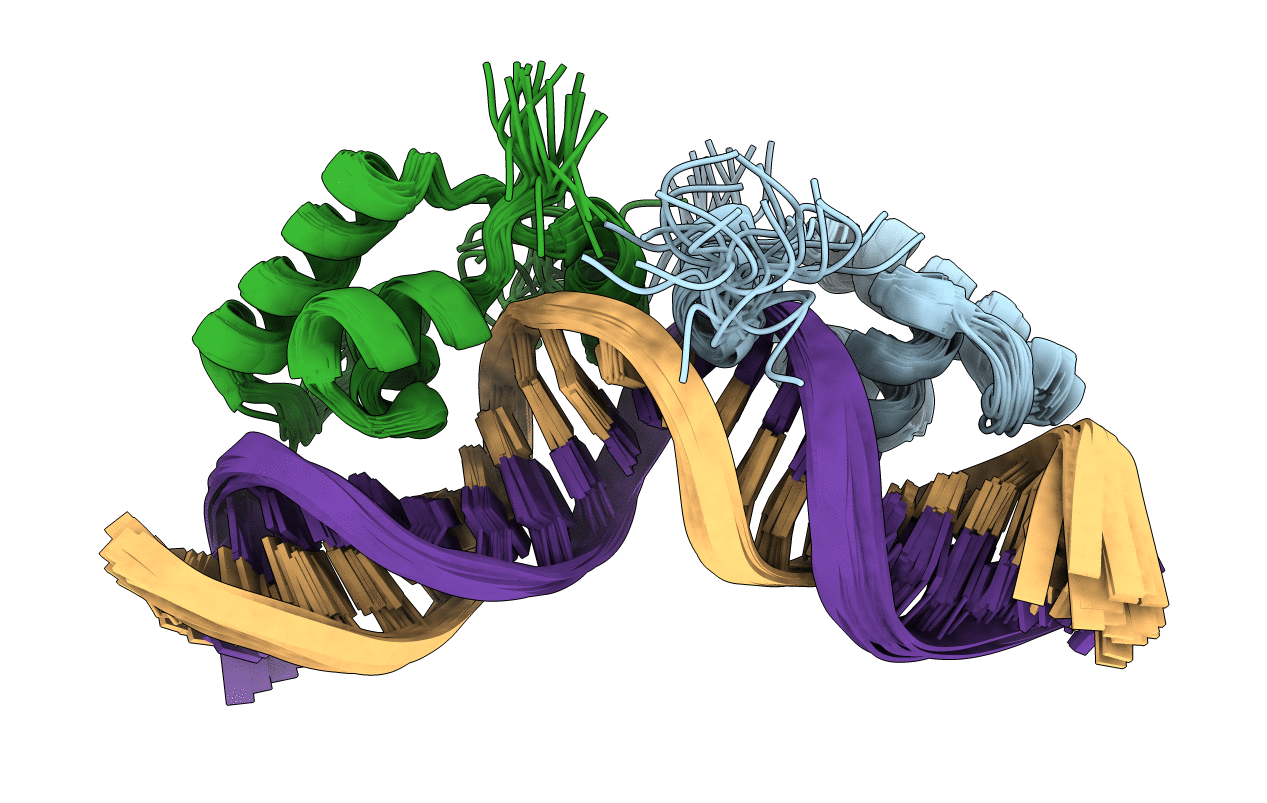
Refined Solution Structure of a Dimer of LAC repressor DNA-Binding domain complexed to its natural operator O1
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 83333)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
200
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy