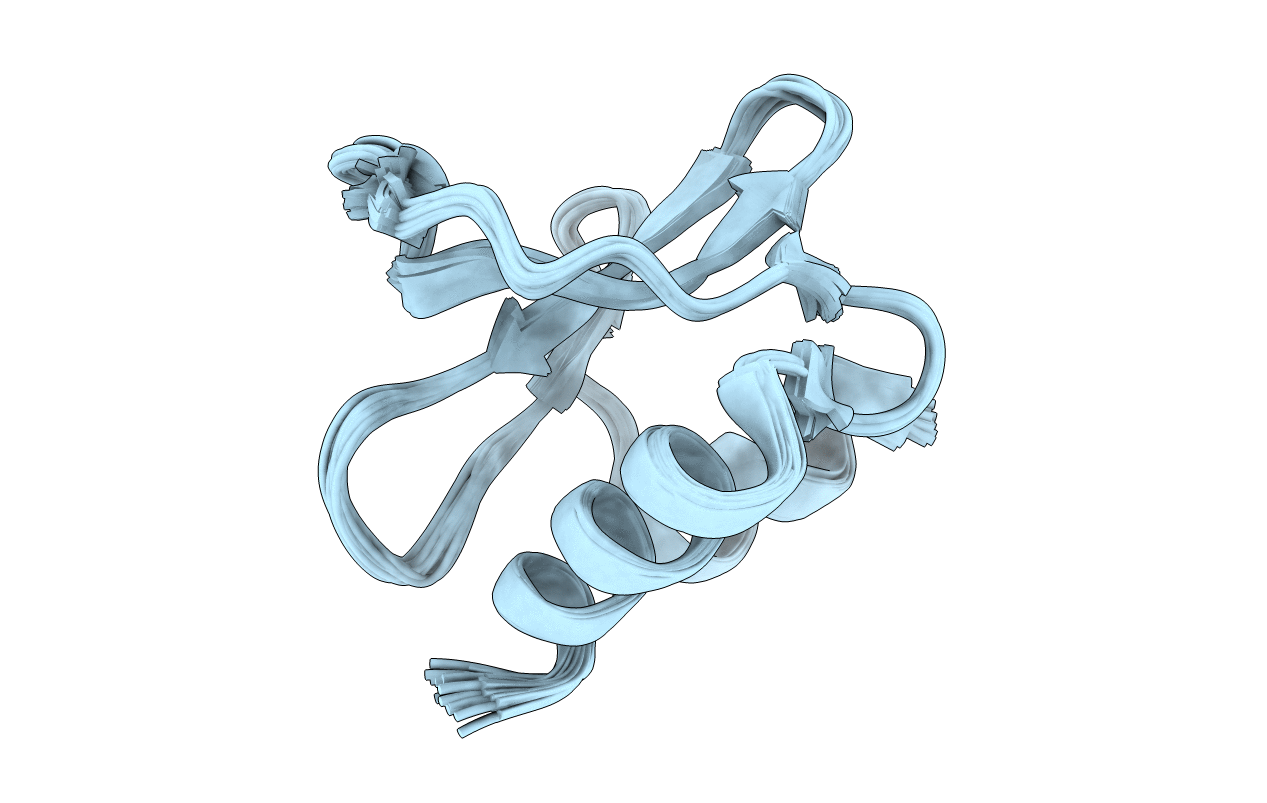
Deposition Date
2008-11-05
Release Date
2008-11-25
Last Version Date
2024-05-01
Entry Detail
PDB ID:
2KAF
Keywords:
Title:
Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus
Biological Source:
Source Organism(s):
SARS coronavirus (Taxon ID: 227859)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
80
Conformers Submitted:
20
Selection Criteria:
target function


