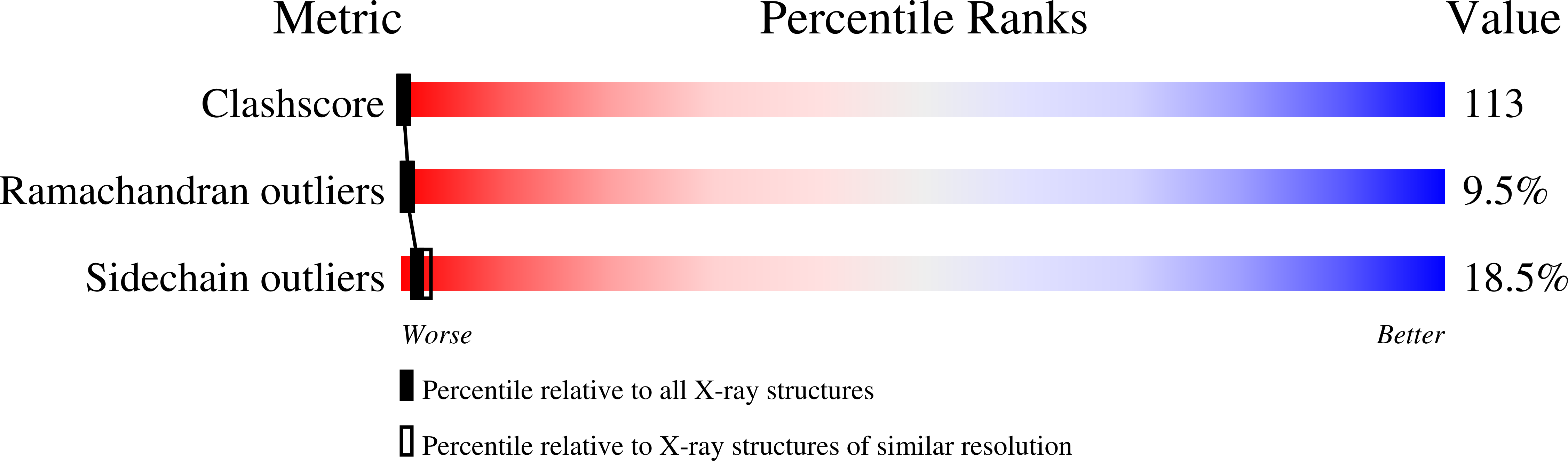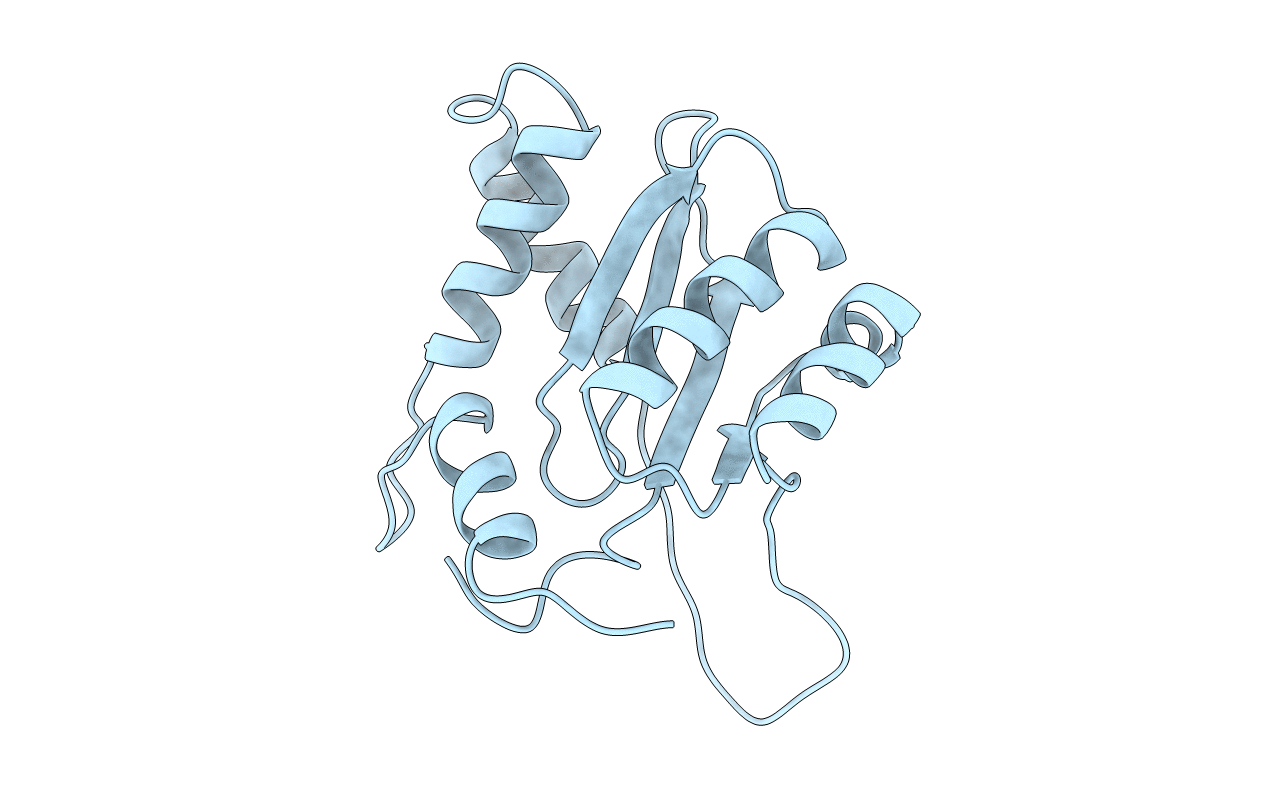
Deposition Date
1996-09-13
Release Date
1997-03-12
Last Version Date
2024-05-29
Entry Detail
PDB ID:
2ITG
Keywords:
Title:
CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT
Biological Source:
Source Organism(s):
Human immunodeficiency virus 1 (Taxon ID: 11676)
Expression System(s):
Method Details: