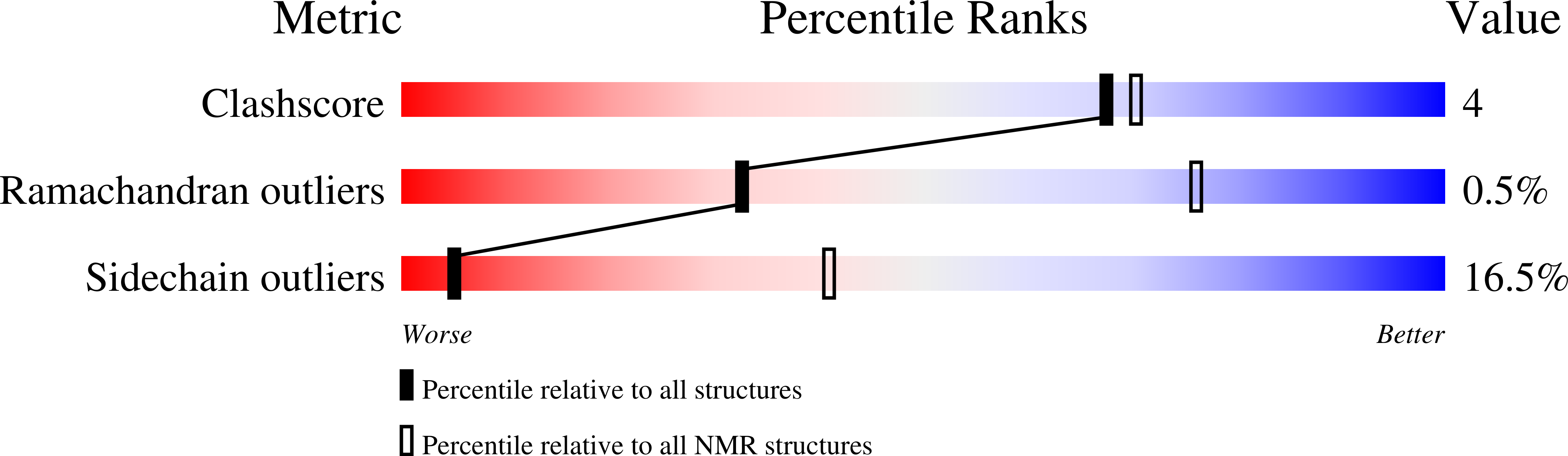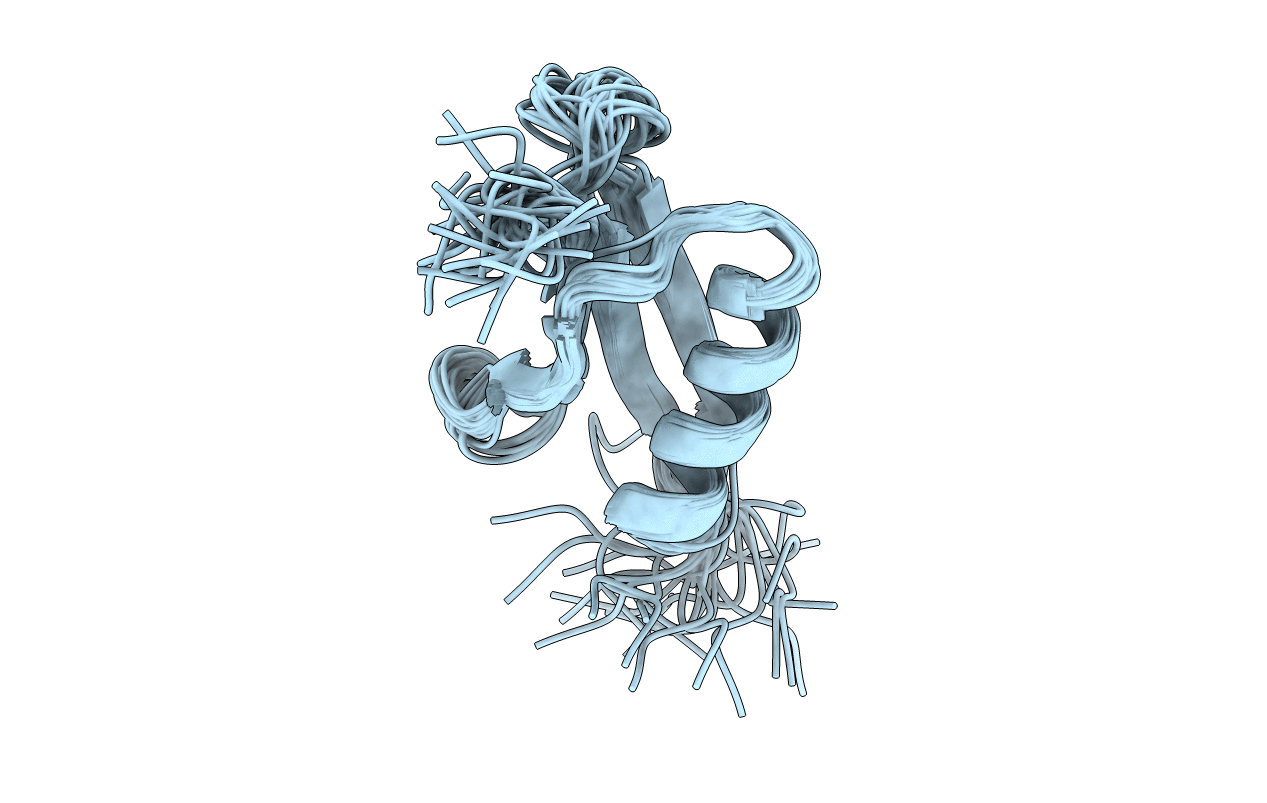
Deposition Date
1992-08-26
Release Date
1994-01-31
Last Version Date
2024-05-29
Entry Detail
PDB ID:
2IGG
Keywords:
Title:
DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR
Biological Source:
Source Organism(s):
Streptococcus sp. GX7805 (Taxon ID: 1325)
Method Details:
Experimental Method:
Conformers Submitted:
27