Deposition Date
2006-08-24
Release Date
2006-09-19
Last Version Date
2024-11-20
Entry Detail
PDB ID:
2I5B
Keywords:
Title:
The crystal structure of an ADP complex of Bacillus subtilis pyridoxal kinase provides evidence for the parralel emergence of enzyme activity during evolution
Biological Source:
Source Organism(s):
Bacillus subtilis (Taxon ID: 1423)
Expression System(s):
Method Details:
Experimental Method:
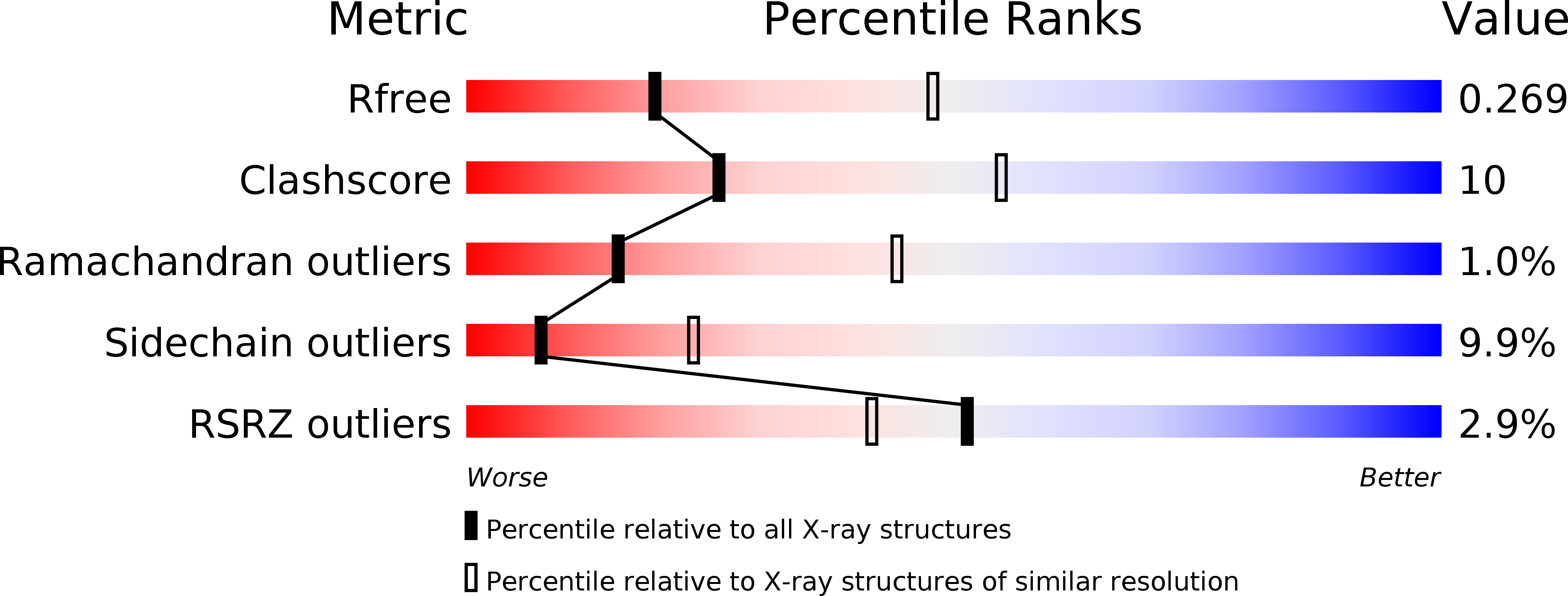
Resolution:
2.80 Å
R-Value Free:
0.27
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 41 21 2