Deposition Date
2006-07-26
Release Date
2007-09-18
Last Version Date
2024-05-08
Entry Detail
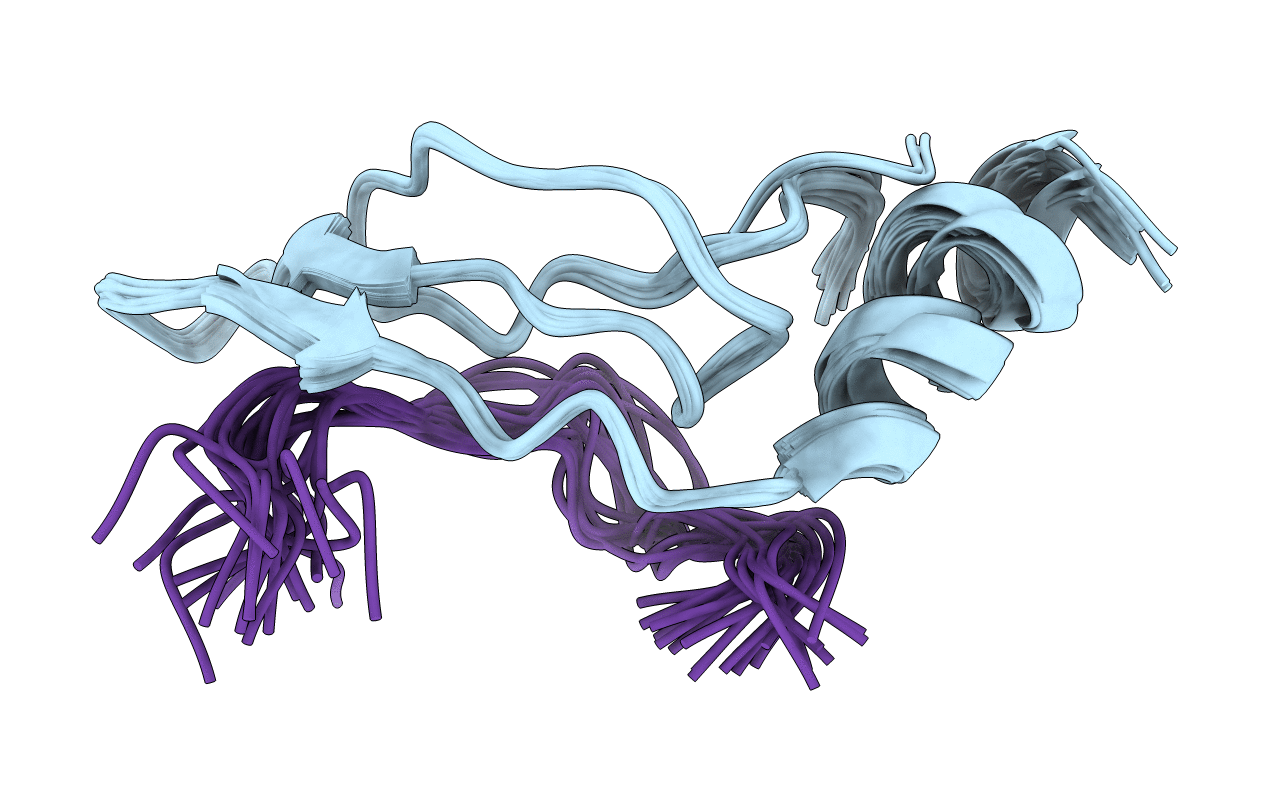
PDB ID:
2HUG
Keywords:
Title:
3D Solution Structure of the Chromo-2 Domain of cpSRP43 complexed with cpSRP54 peptide
Biological Source:
Source Organism(s):
Arabidopsis thaliana (Taxon ID: 3702)
Expression System(s):
Method Details:


