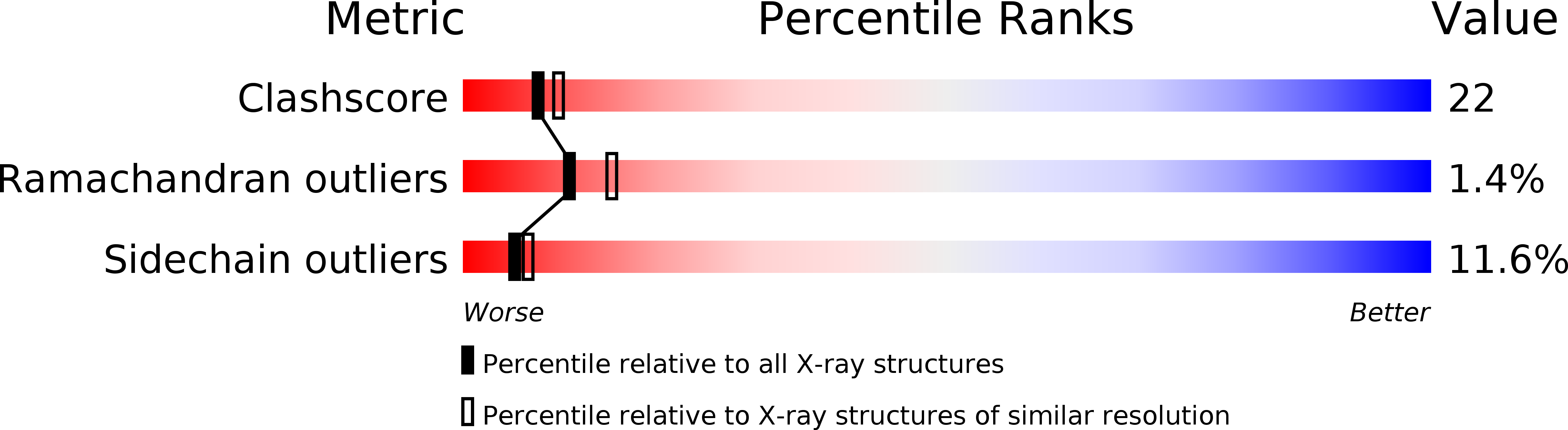
Deposition Date
1994-03-28
Release Date
1994-08-31
Last Version Date
2024-02-14
Entry Detail
PDB ID:
2HSD
Keywords:
Title:
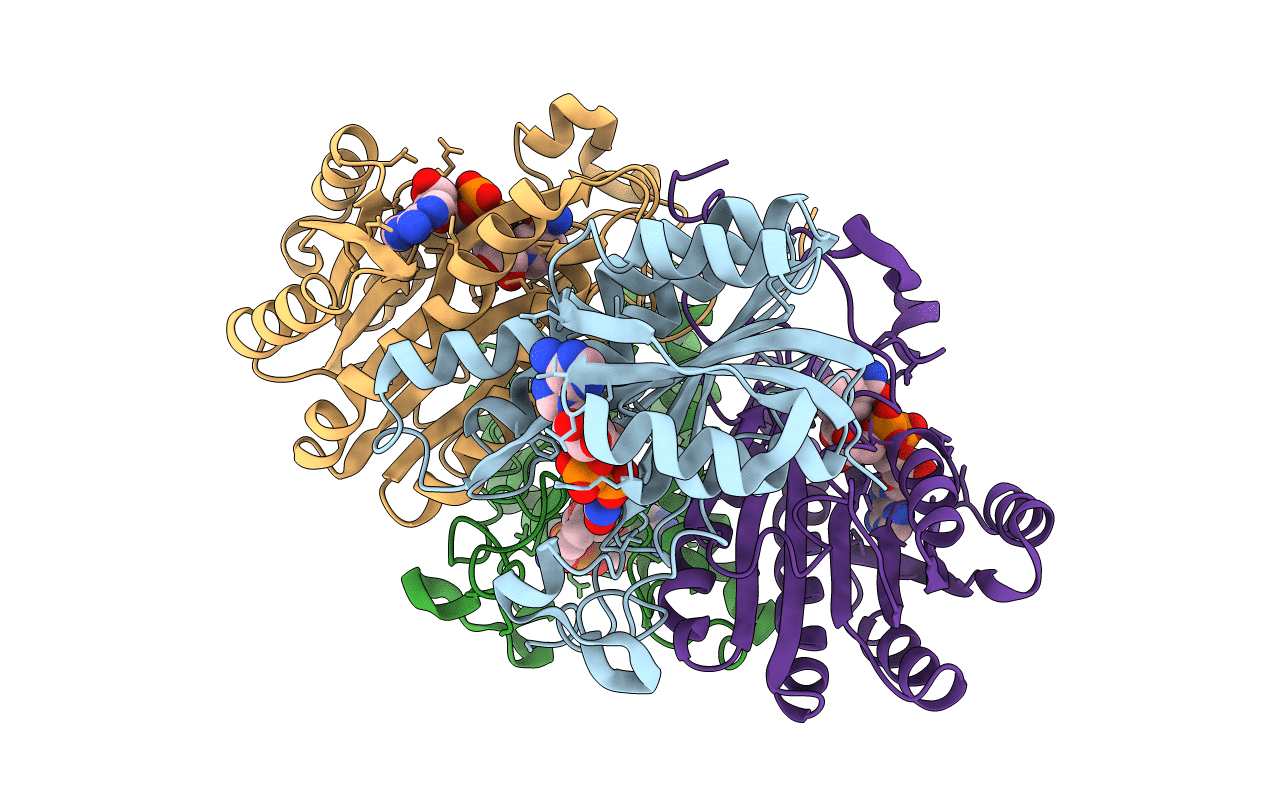
THE REFINED THREE-DIMENSIONAL STRUCTURE OF 3ALPHA,20BETA-HYDROXYSTEROID DEHYDROGENASE AND POSSIBLE ROLES OF THE RESIDUES CONSERVED IN SHORT-CHAIN DEHYDROGENASES
Biological Source:
Source Organism(s):
Streptomyces exfoliatus (Taxon ID: 1905)
Method Details:
Experimental Method:
Resolution:
2.64 Å
R-Value Observed:
0.18
Space Group:
P 43 21 2