Deposition Date
2006-06-11
Release Date
2006-12-12
Last Version Date
2024-02-14
Entry Detail
PDB ID:
2H9S
Keywords:
Title:
Crystal Structure of Homo-DNA and Nature's Choice of Pentose over Hexose in the Genetic System
Method Details:
Experimental Method:
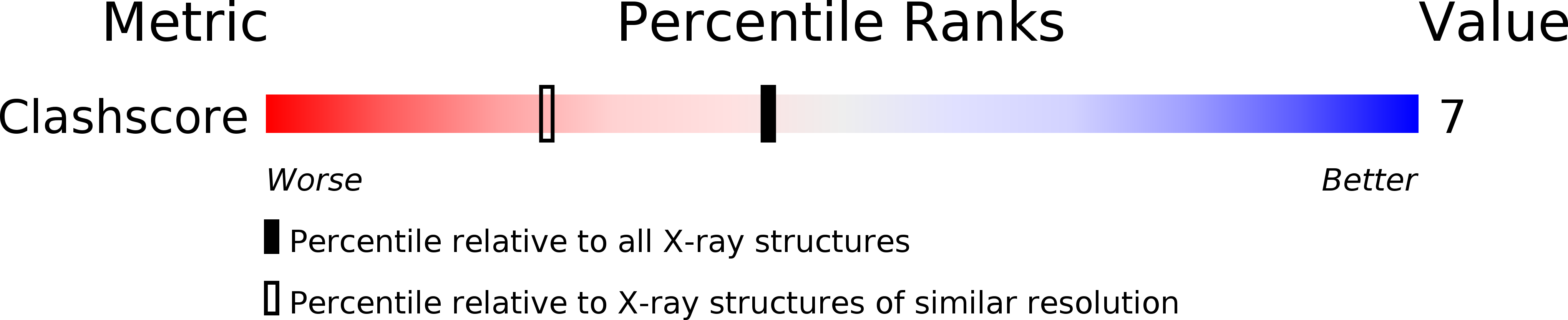
Resolution:
1.75 Å
R-Value Free:
0.28
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 61 2 2