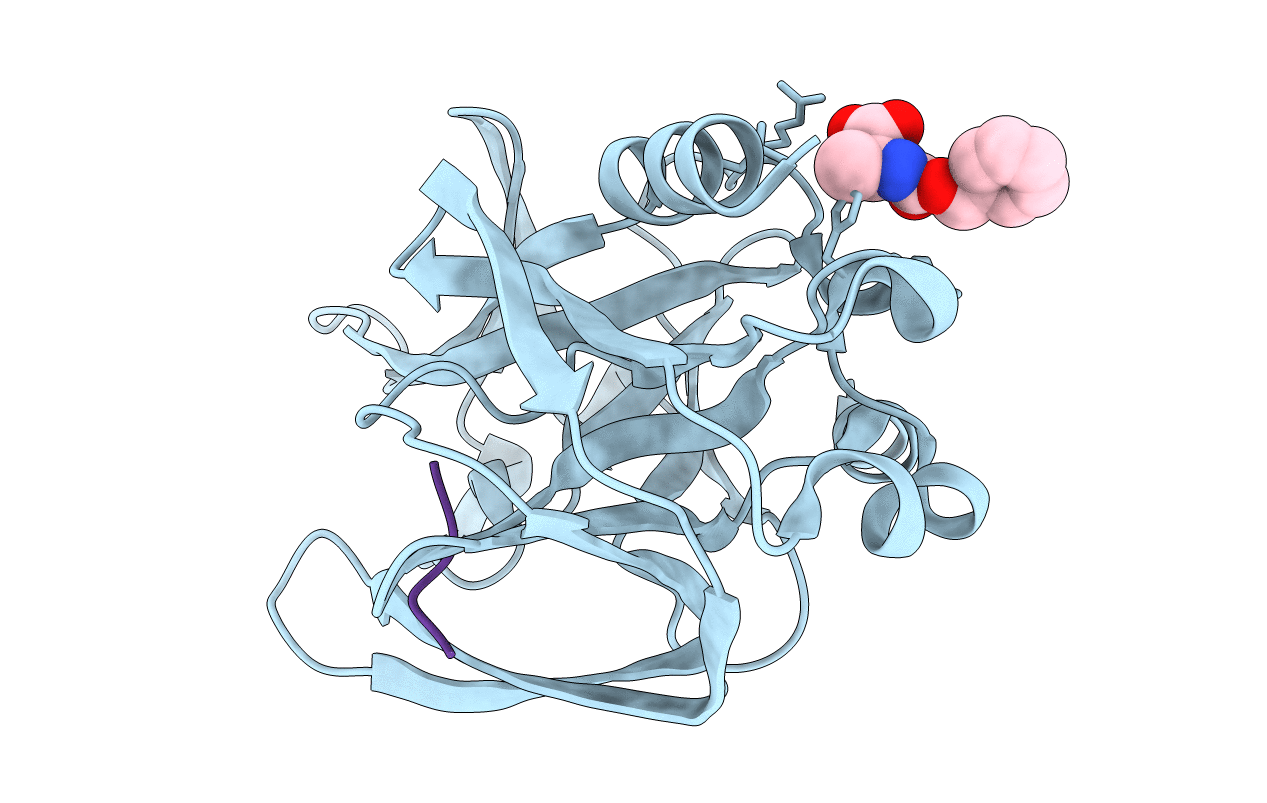
Deposition Date
2006-05-31
Release Date
2006-08-08
Last Version Date
2021-10-20
Entry Detail
PDB ID:
2H6M
Keywords:
Title:
An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors
Biological Source:
Source Organism(s):
Hepatitis A virus (Taxon ID: 12092)
Expression System(s):
Method Details:
Experimental Method:
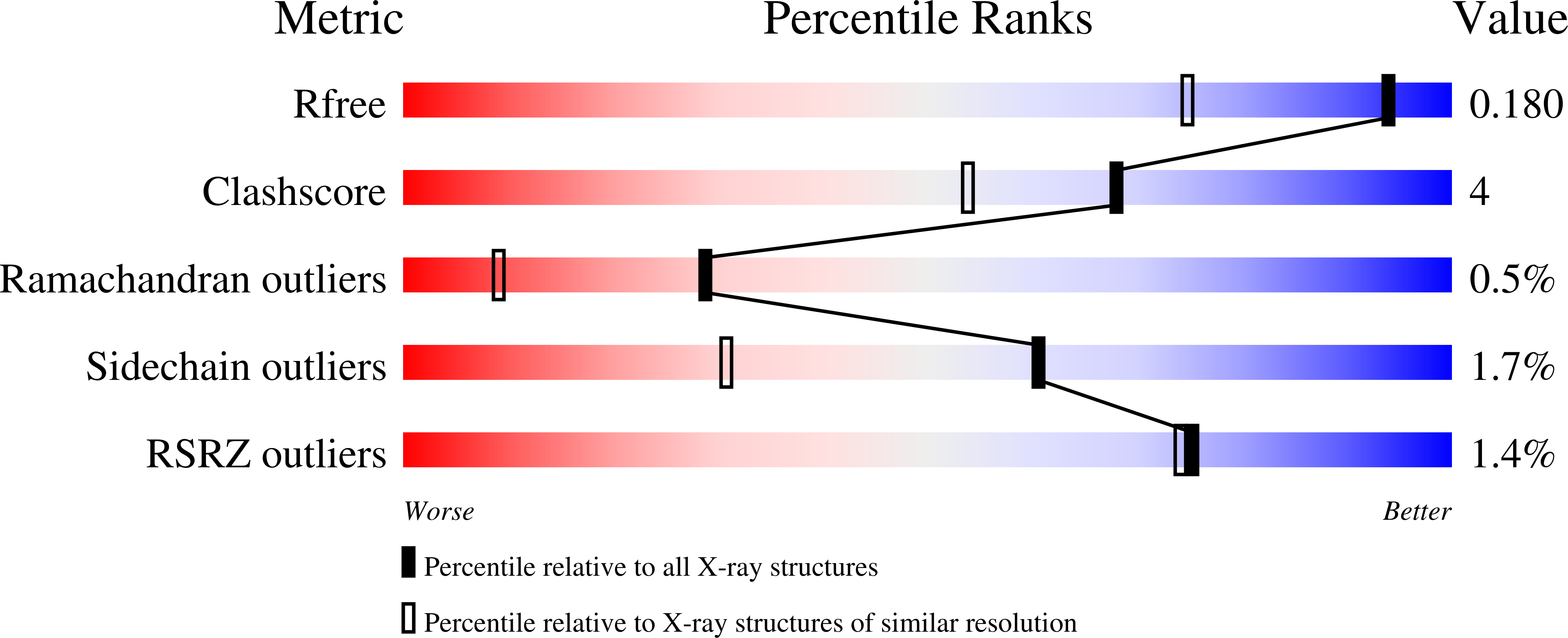
Resolution:
1.40 Å
R-Value Free:
0.20
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 21 21 21