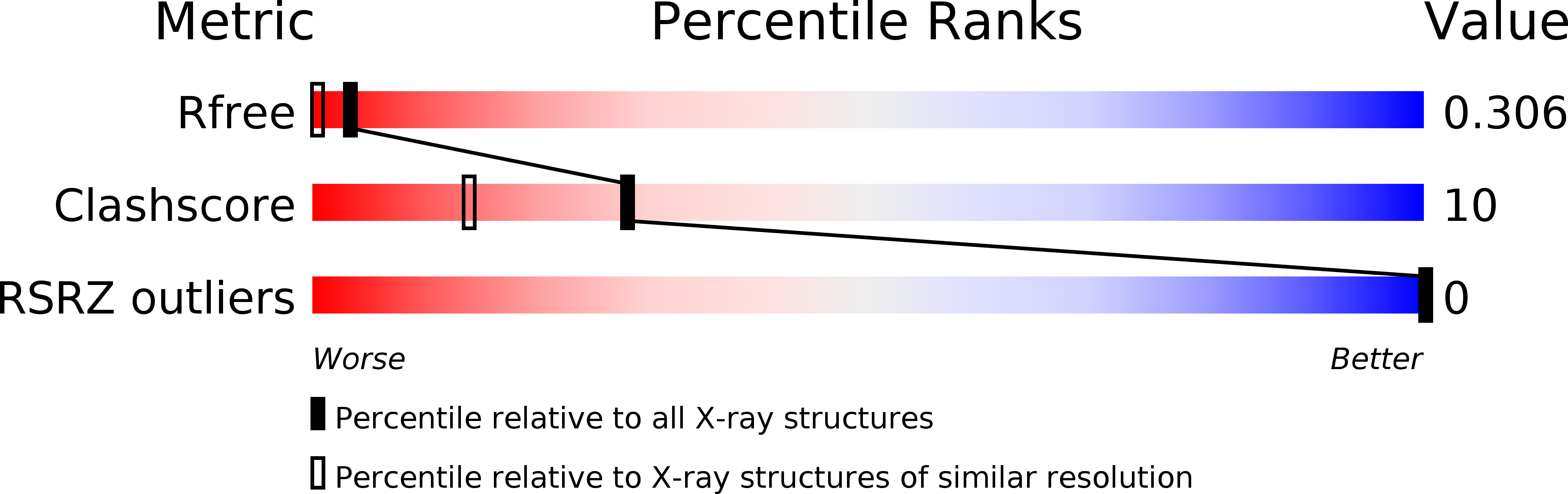Deposition Date
2006-05-10
Release Date
2007-05-22
Last Version Date
2023-08-30
Entry Detail
PDB ID:
2GYX
Keywords:
Title:
Crystal structure of DB884- D(CGCGAATTCGCG)2 complex at 1.86 A resolution.
Method Details:
Experimental Method:
Resolution:
1.86 Å
R-Value Free:
0.32
R-Value Observed:
0.23
Space Group:
P 21 21 21