Deposition Date
2006-05-09
Release Date
2007-03-20
Last Version Date
2024-11-06
Entry Detail
PDB ID:
2GYP
Keywords:
Title:
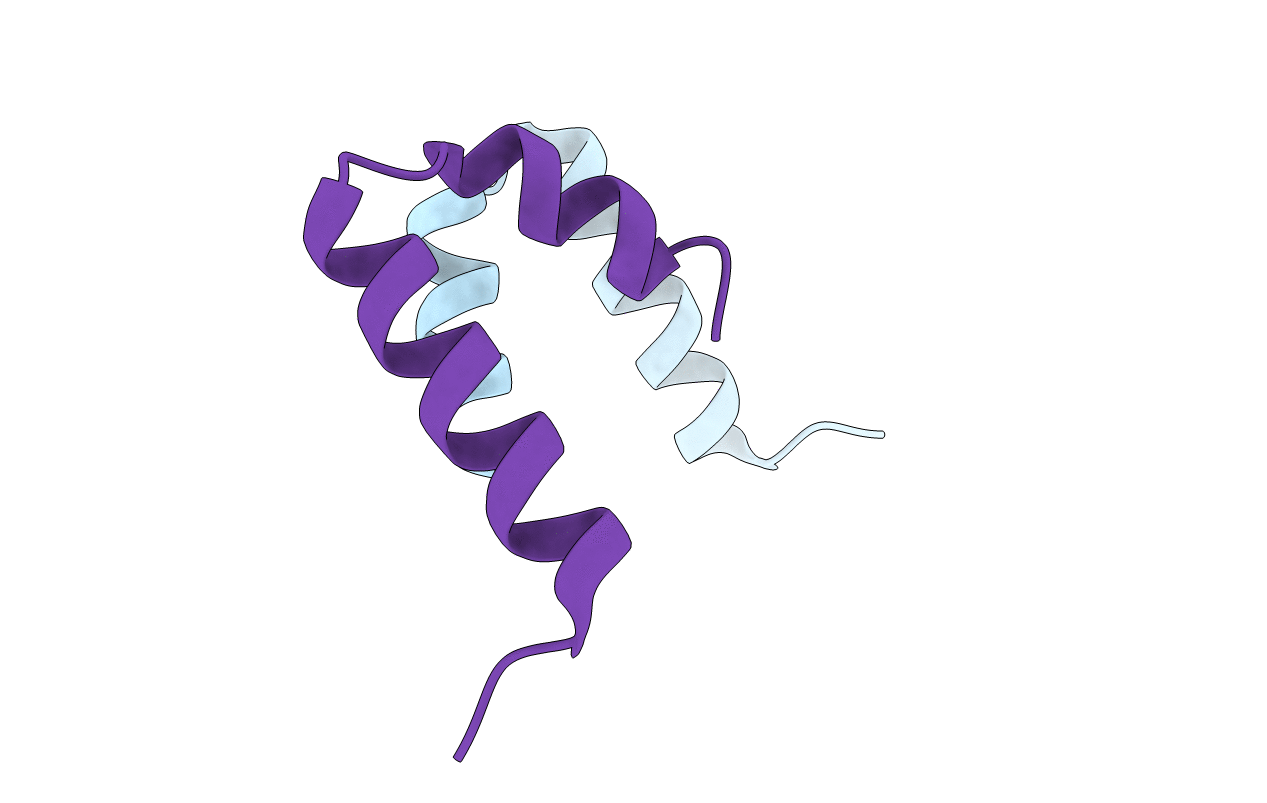
Diabetes mellitus due to a frustrated Schellman motif in HNF-1a
Method Details:
Experimental Method:
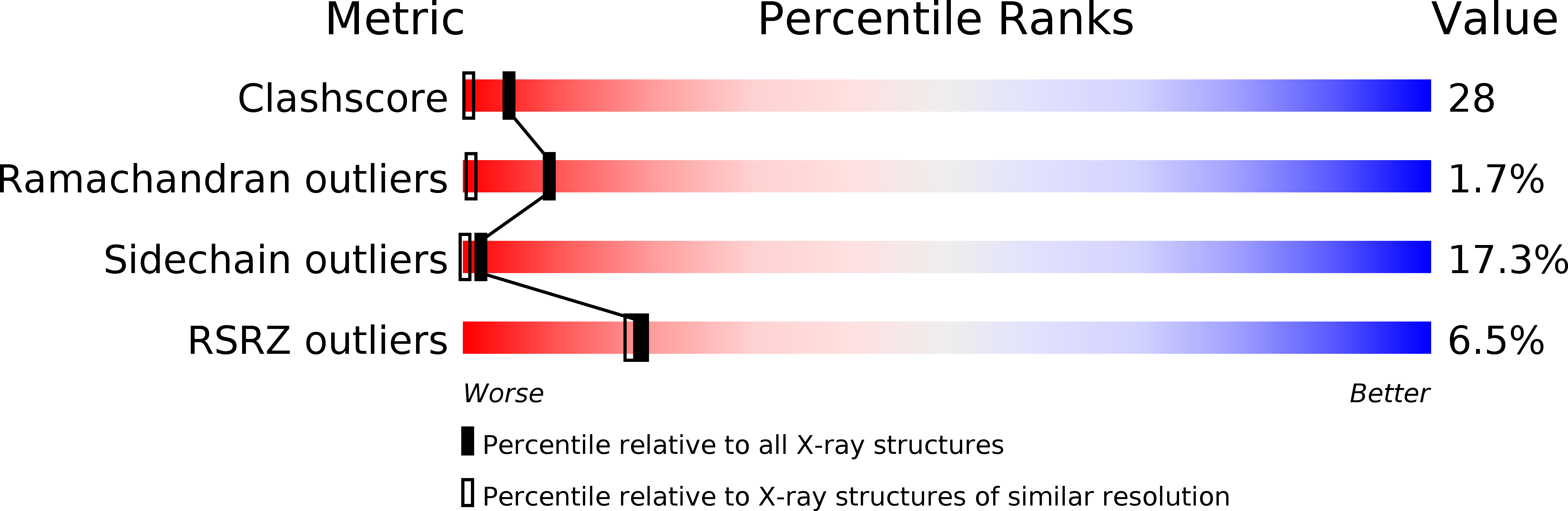
Resolution:
1.40 Å
R-Value Free:
0.28
R-Value Work:
0.22
R-Value Observed:
0.23
Space Group:
P 21 21 2