Deposition Date
2006-05-02
Release Date
2006-07-04
Last Version Date
2024-10-30
Entry Detail
PDB ID:
2GVD
Keywords:
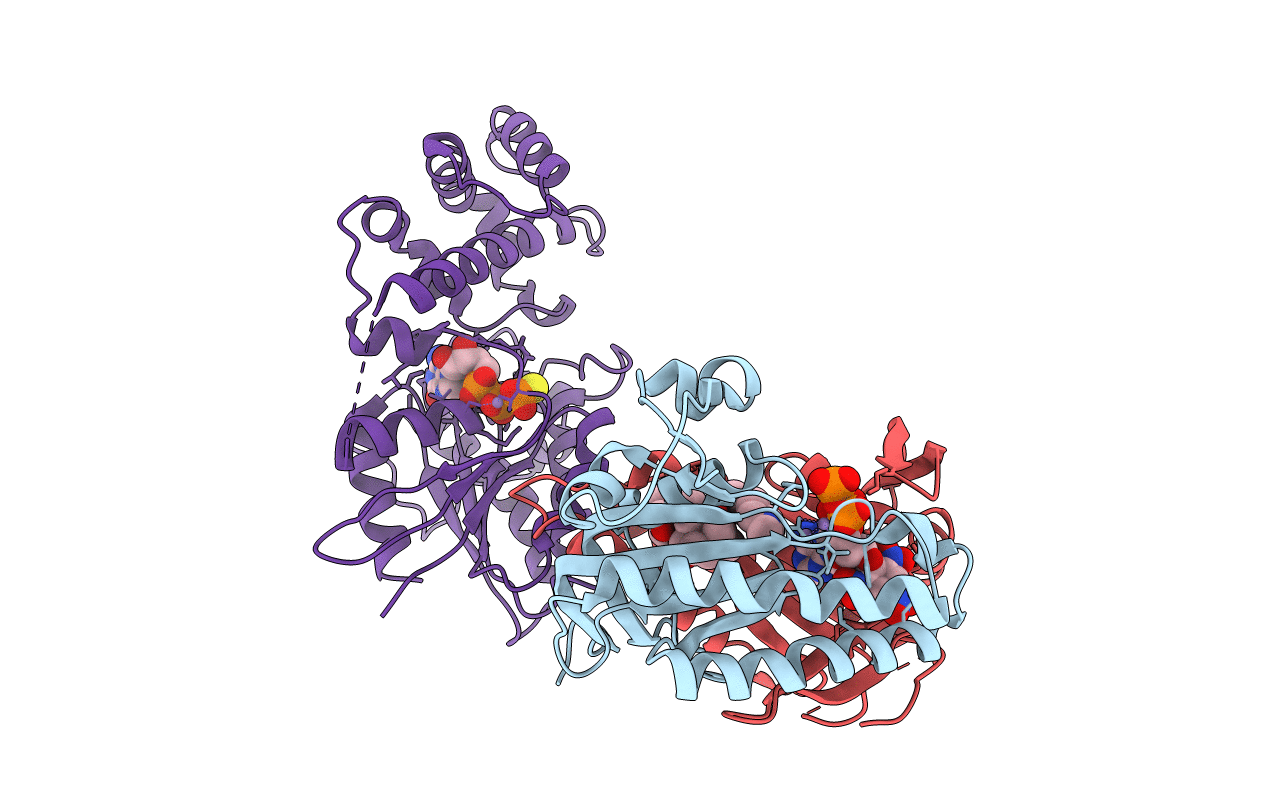
Title:
Complex Of Gs- With The Catalytic Domains Of Mammalian Adenylyl Cyclase: Complex With TNP-ATP and Mn
Biological Source:
Source Organism(s):
Canis lupus familiaris (Taxon ID: 9615)
Rattus norvegicus (Taxon ID: 10116)
Bos taurus (Taxon ID: 9913)
Rattus norvegicus (Taxon ID: 10116)
Bos taurus (Taxon ID: 9913)
Expression System(s):
Method Details:
Experimental Method:
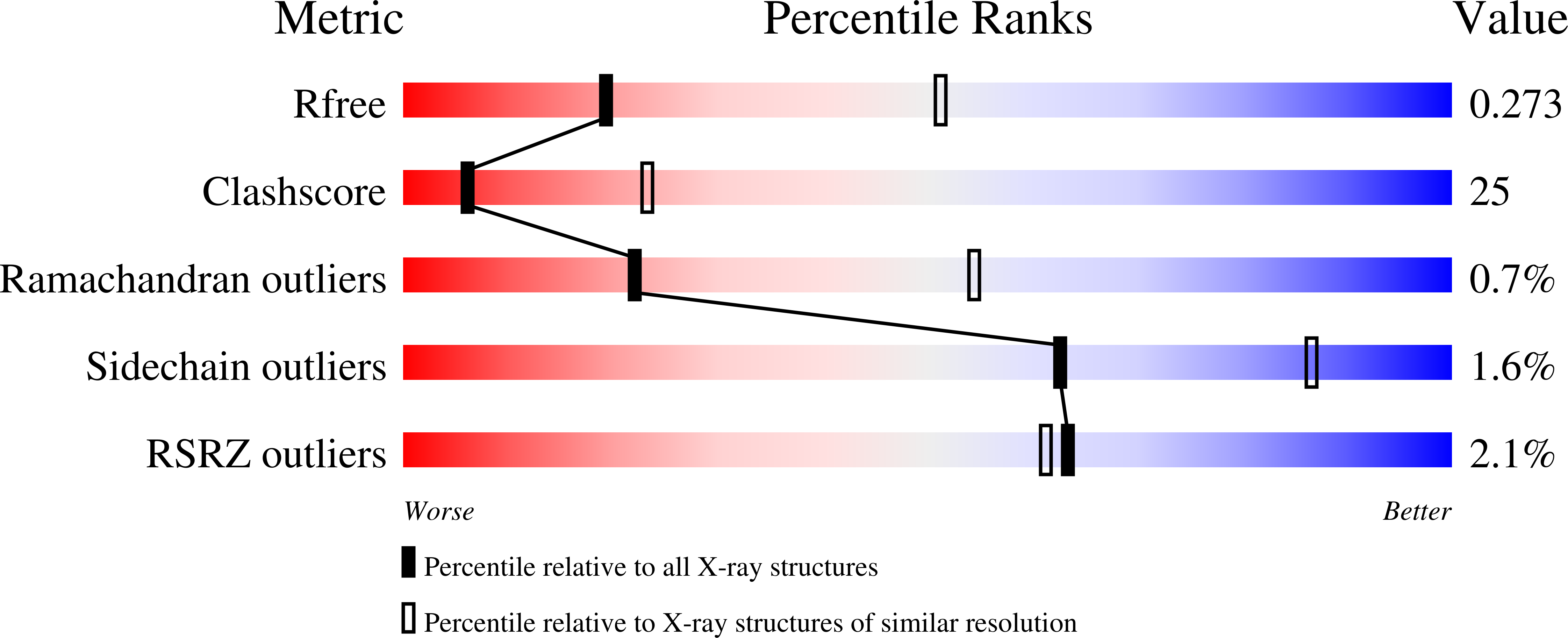
Resolution:
2.90 Å
R-Value Free:
0.27
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 21 21 2