Deposition Date
2006-04-17
Release Date
2006-09-12
Last Version Date
2024-10-16
Entry Detail
PDB ID:
2GP9
Keywords:
Title:
Crystal structure of the slow form of thrombin in a self-inhibited conformation
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
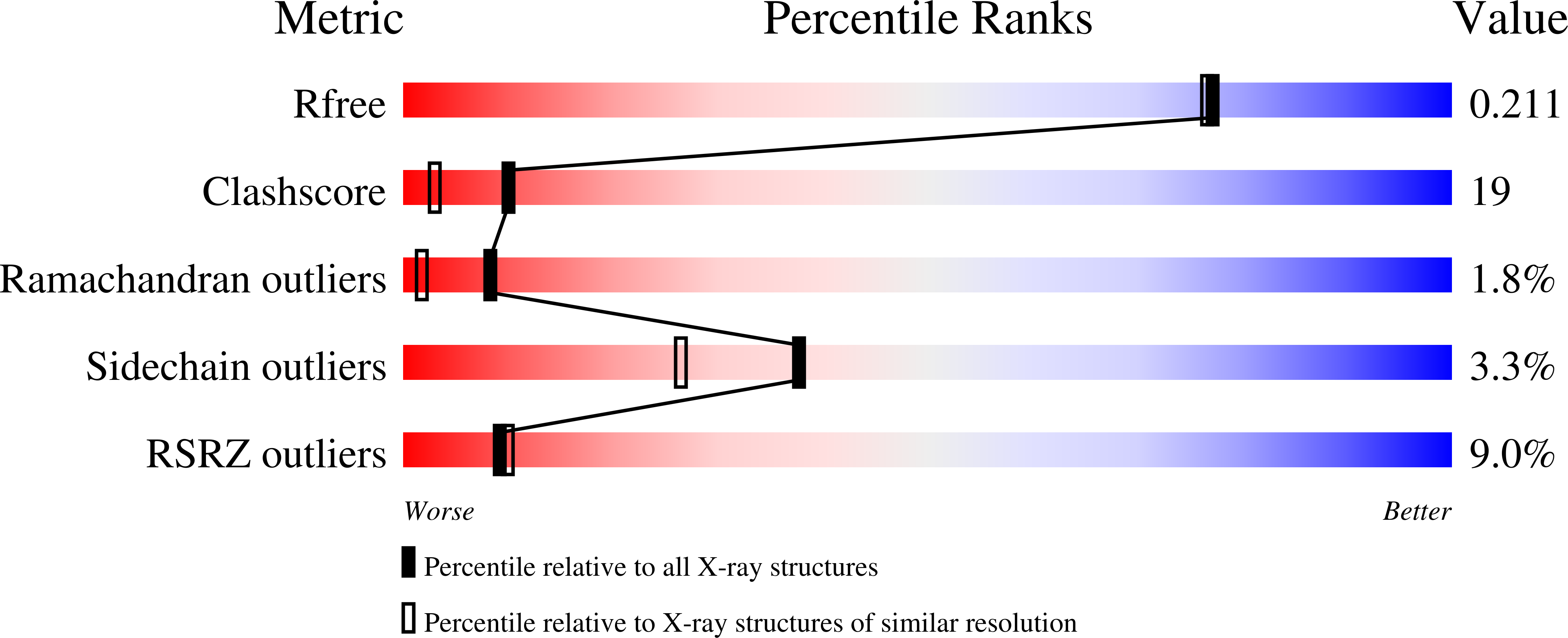
Resolution:
1.87 Å
R-Value Free:
0.21
R-Value Work:
0.19
Space Group:
P 43