Deposition Date
2006-03-04
Release Date
2006-04-18
Last Version Date
2024-04-03
Entry Detail
PDB ID:
2G92
Keywords:
Title:
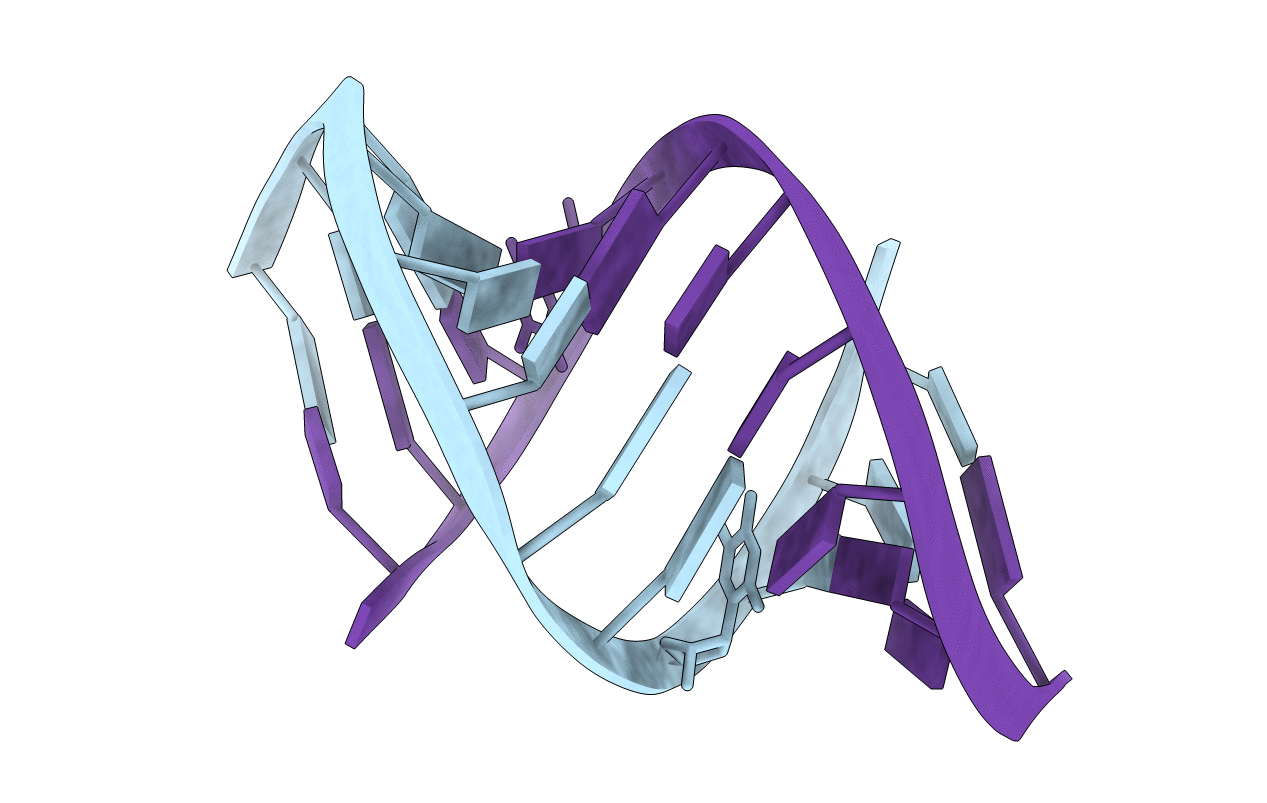
Crystal Structure Analysis of the RNA Dodecamer CGC-(NF2)-AAUUAGCG, with an Incorporated 2,4-Difluorotoluyl Residue (NF2)
Method Details:
Experimental Method:
Resolution:
1.61 Å
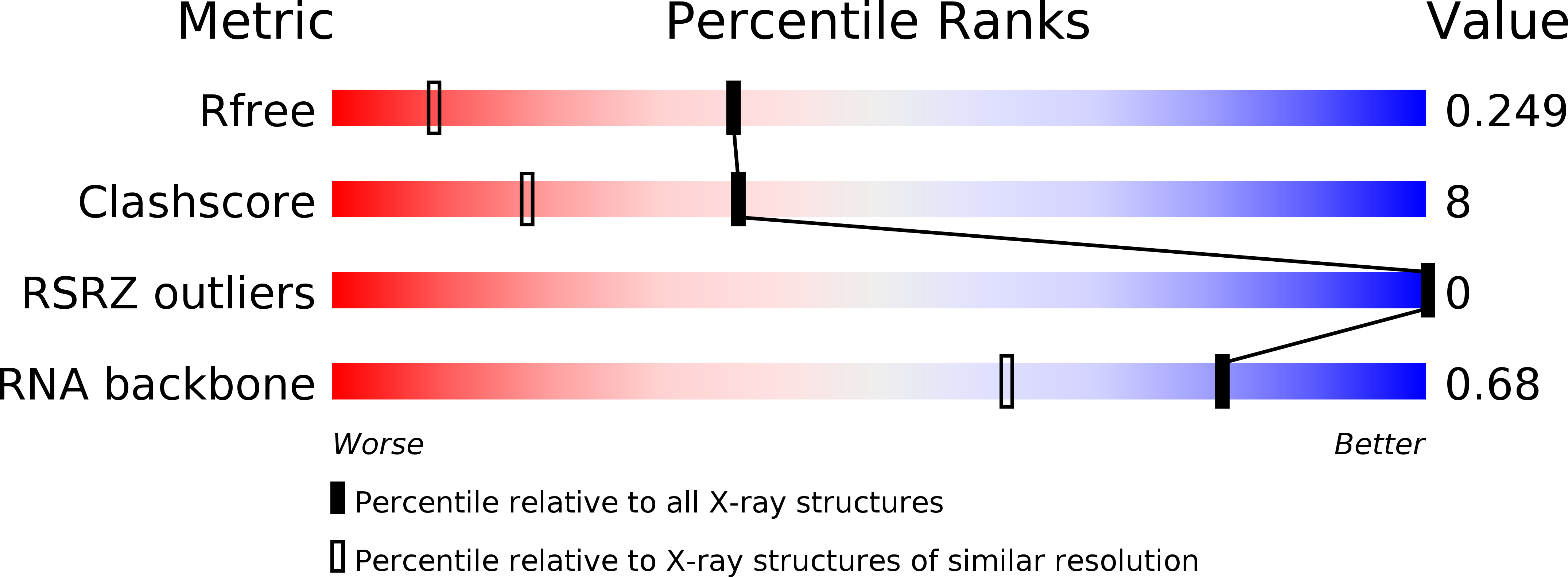
R-Value Free:
0.23
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 31