Deposition Date
2006-02-16
Release Date
2006-03-14
Last Version Date
2024-11-20
Entry Detail
PDB ID:
2G2X
Keywords:
Title:
X-Ray Crystal Structure Protein Q88CH6 from Pseudomonas putida. Northeast Structural Genomics Consortium Target PpR72.
Biological Source:
Source Organism(s):
Pseudomonas putida (Taxon ID: 160488)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.30 Å
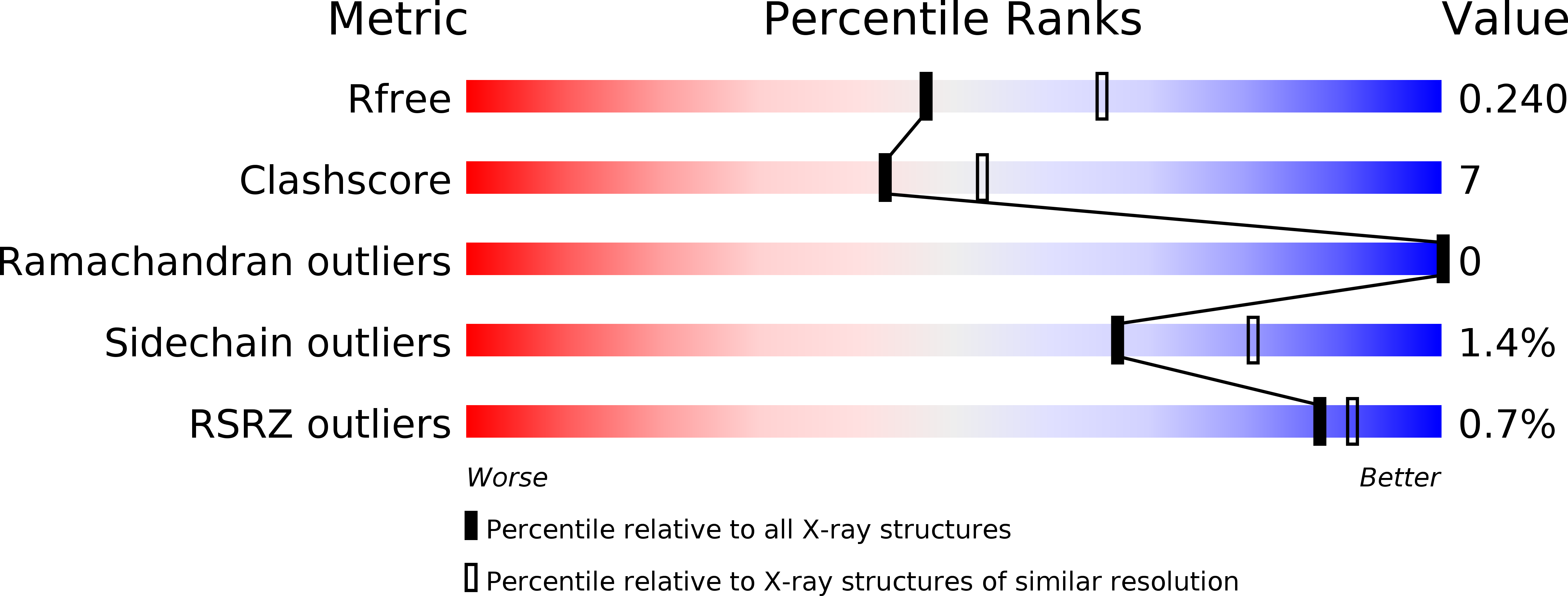
R-Value Free:
0.24
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
C 1 2 1