Deposition Date
2005-11-10
Release Date
2006-04-25
Last Version Date
2023-11-15
Entry Detail
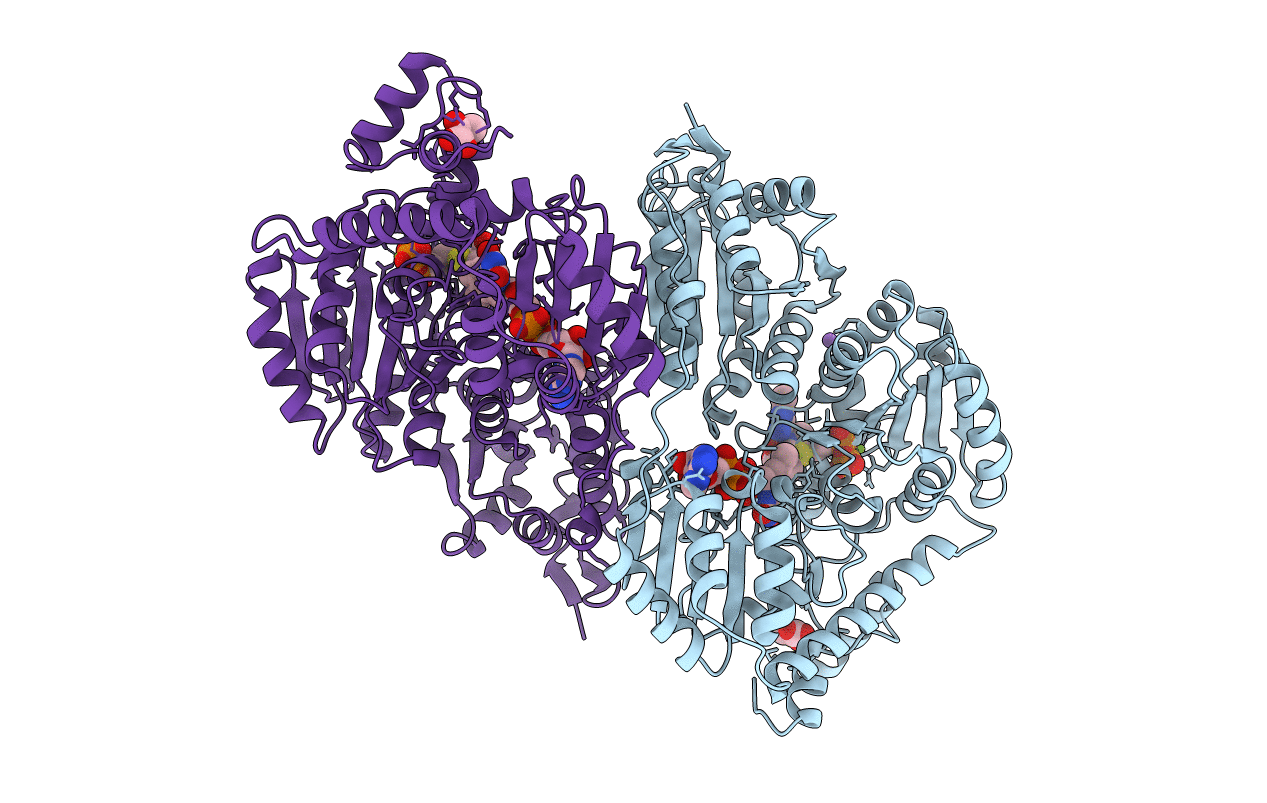
PDB ID:
2EZ8
Keywords:
Title:
Pyruvate oxidase variant F479W in complex with reaction intermediate 2-lactyl-thiamin diphosphate
Biological Source:
Source Organism(s):
Lactobacillus plantarum (Taxon ID: 1590)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.96 Å
R-Value Free:
0.17
R-Value Work:
0.14
R-Value Observed:
0.14
Space Group:
C 2 2 21


