Deposition Date
2005-11-09
Release Date
2006-03-21
Last Version Date
2024-11-13
Entry Detail
PDB ID:
2EYT
Keywords:
Title:
A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
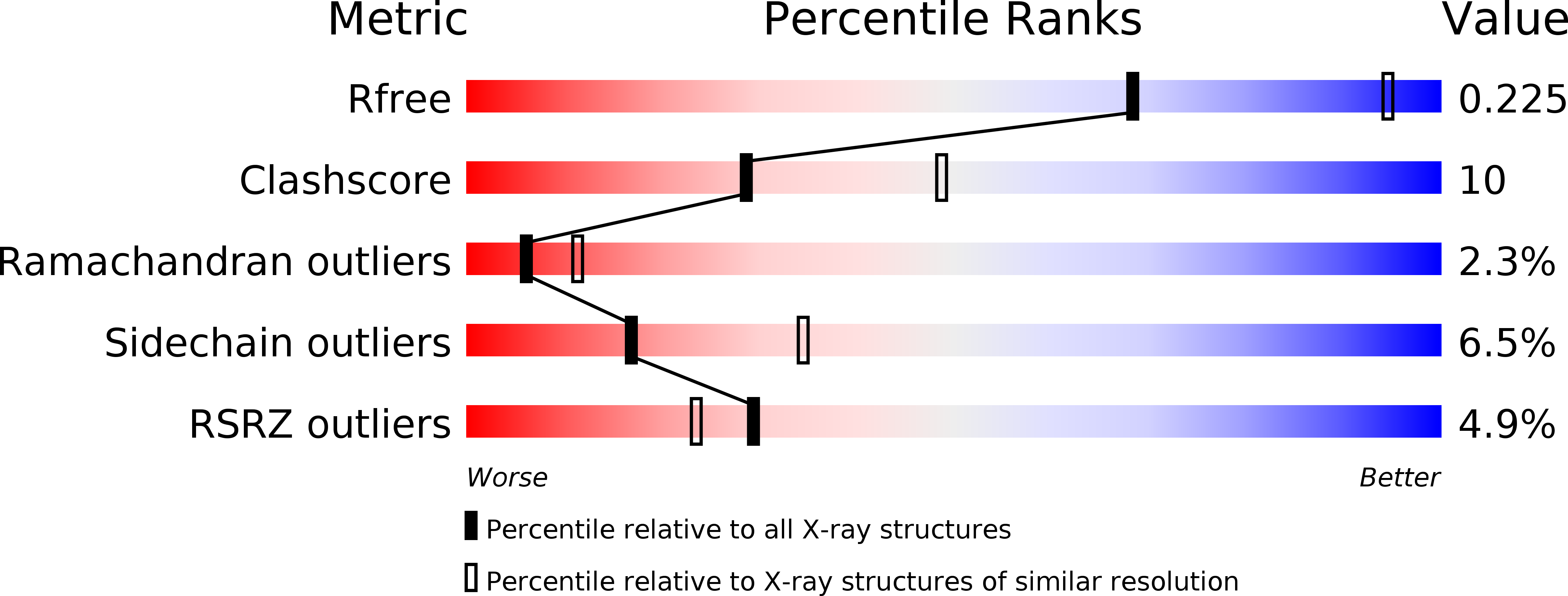
Resolution:
2.60 Å
R-Value Free:
0.25
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 32