Deposition Date
2005-09-12
Release Date
2005-10-25
Last Version Date
2024-10-23
Entry Detail
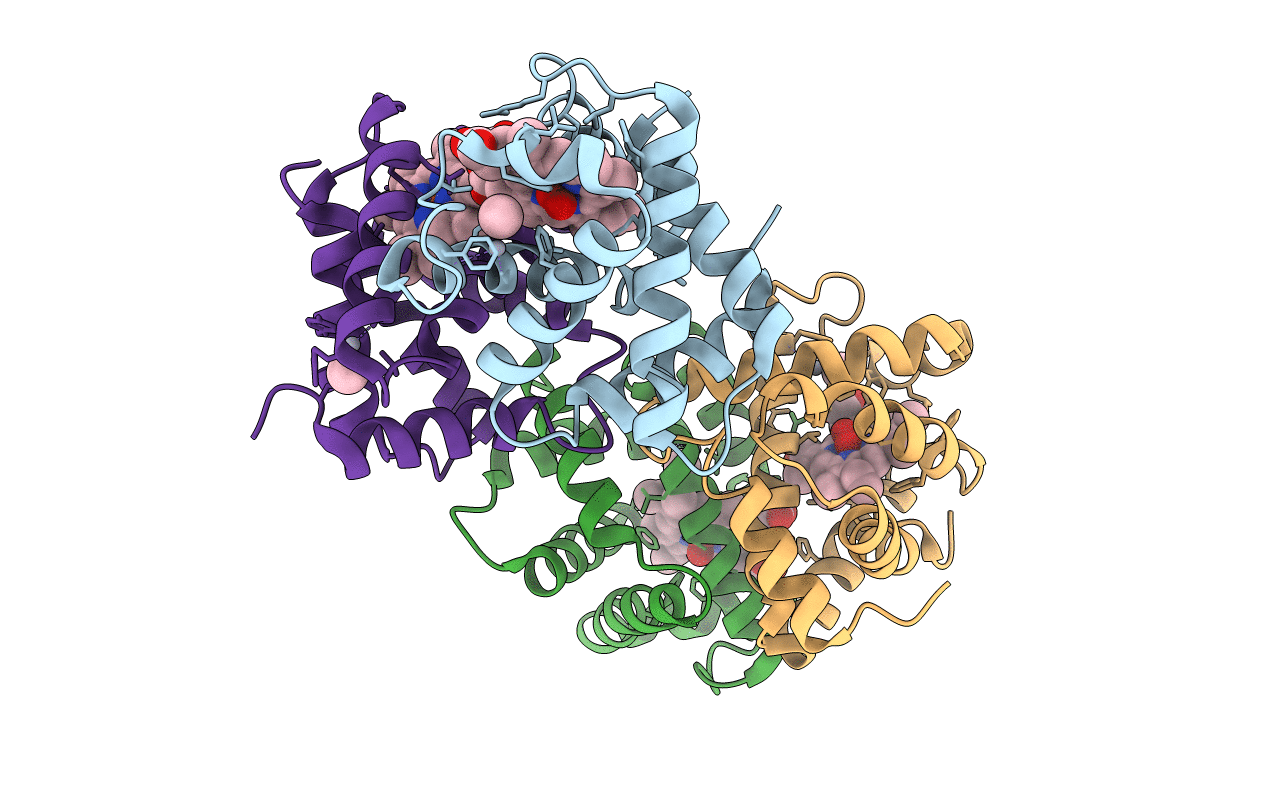
PDB ID:
2D2N
Keywords:
Title:
Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi
Biological Source:
Source Organism(s):
Oligobrachia mashikoi (Taxon ID: 55676)
Method Details:
Experimental Method:
Resolution:
3.20 Å
R-Value Free:
0.29
R-Value Work:
0.24
Space Group:
H 3 2


