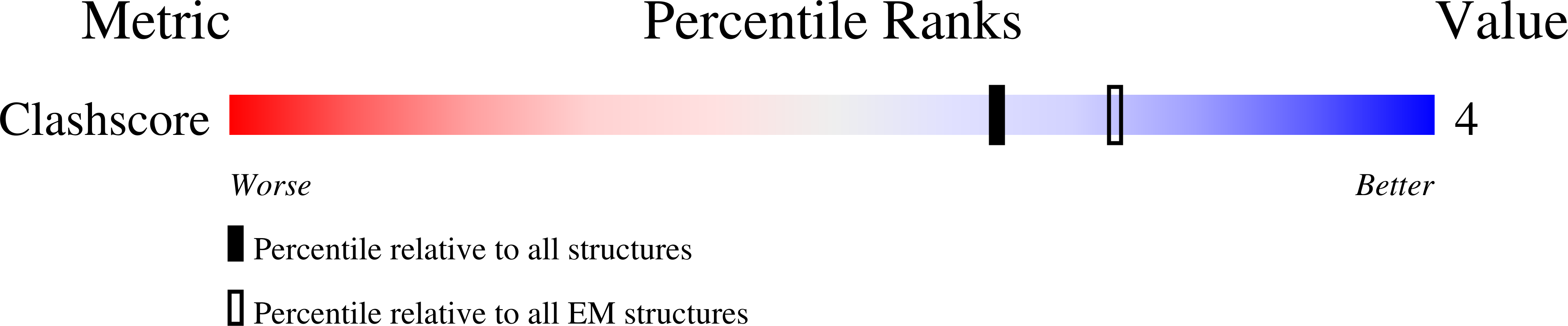
Deposition Date
2005-12-05
Release Date
2006-01-17
Last Version Date
2024-05-08
Entry Detail
PDB ID:
2C8I
Keywords:
Title:
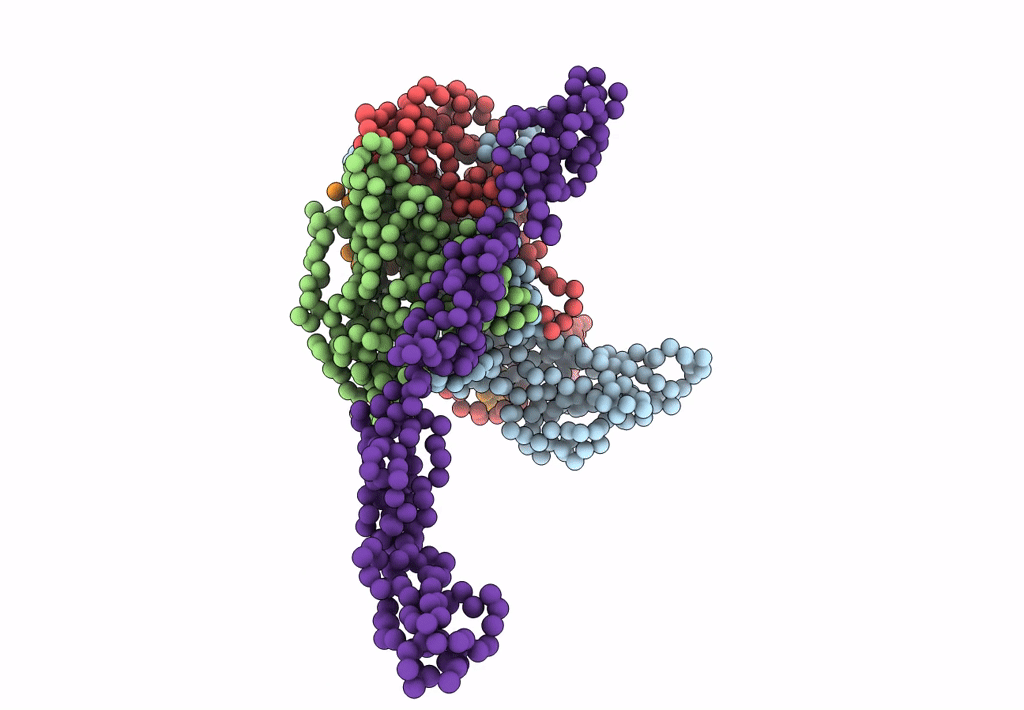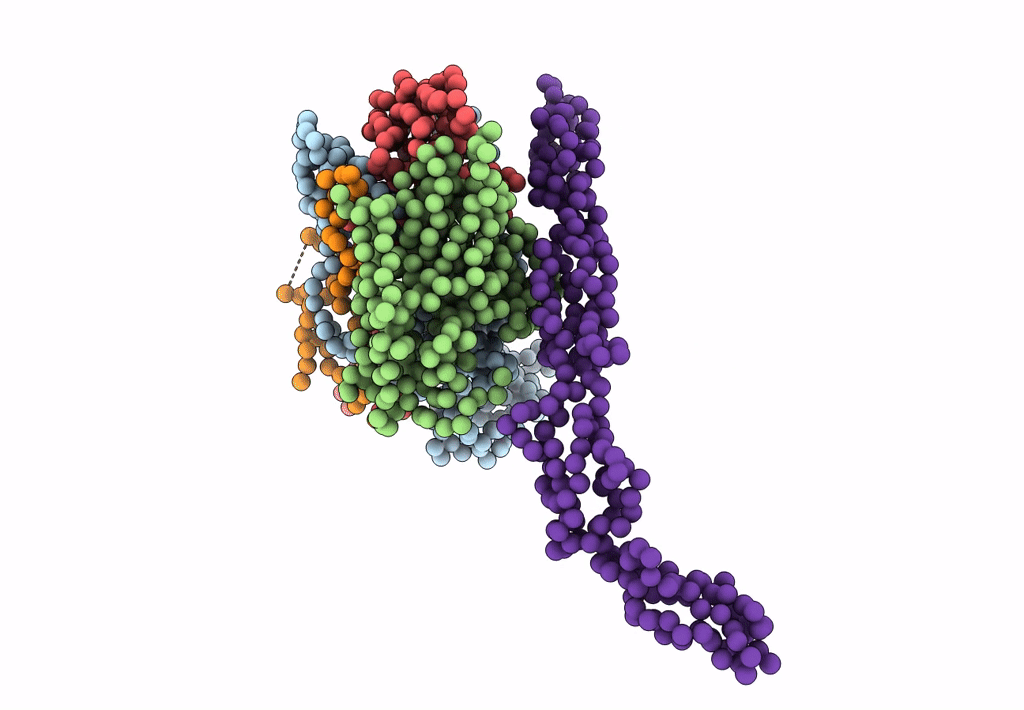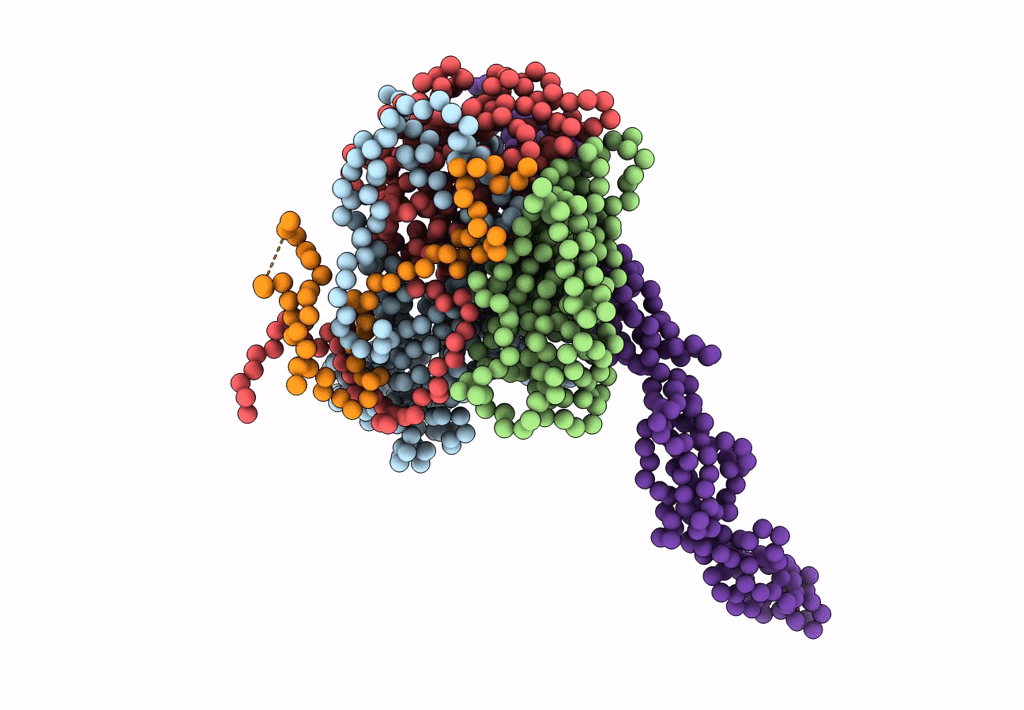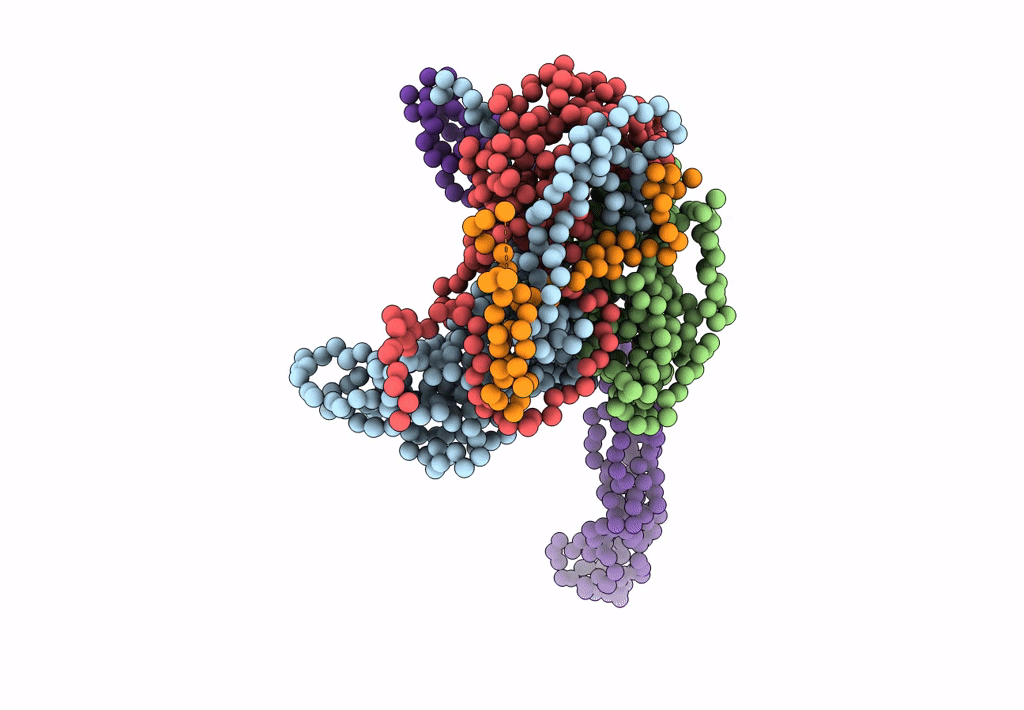
Complex Of Echovirus Type 12 With Domains 1, 2, 3 and 4 Of Its Receptor Decay Accelerating Factor (Cd55) By Cryo Electron Microscopy At 16 A
Biological Source:
Source Organism(s):
HOMO SAPIENS (Taxon ID: 9606)
HUMAN ECHOVIRUS 11 (Taxon ID: 12078)
HUMAN ECHOVIRUS 11 (Taxon ID: 12078)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
14.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE