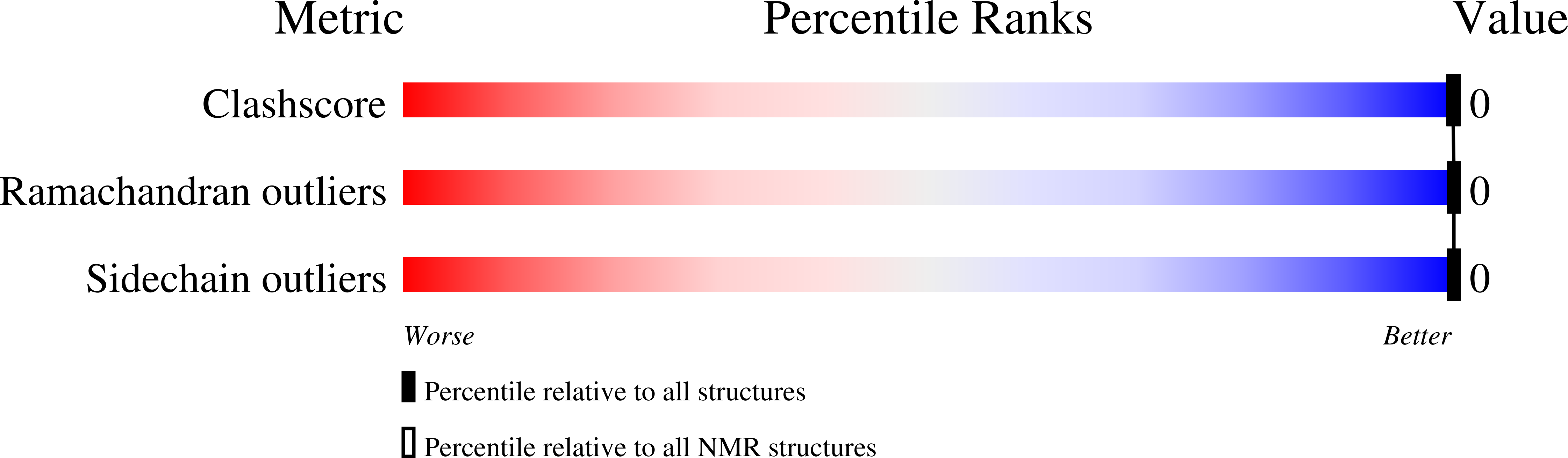
Deposition Date
2005-09-29
Release Date
2006-08-29
Last Version Date
2022-03-09
Entry Detail
Method Details:
Experimental Method:
Conformers Calculated:
211
Conformers Submitted:
16
Selection Criteria:
structures with the lowest energy