Deposition Date
2005-09-15
Release Date
2005-10-04
Last Version Date
2024-11-20
Entry Detail
PDB ID:
2B17
Keywords:
Title:
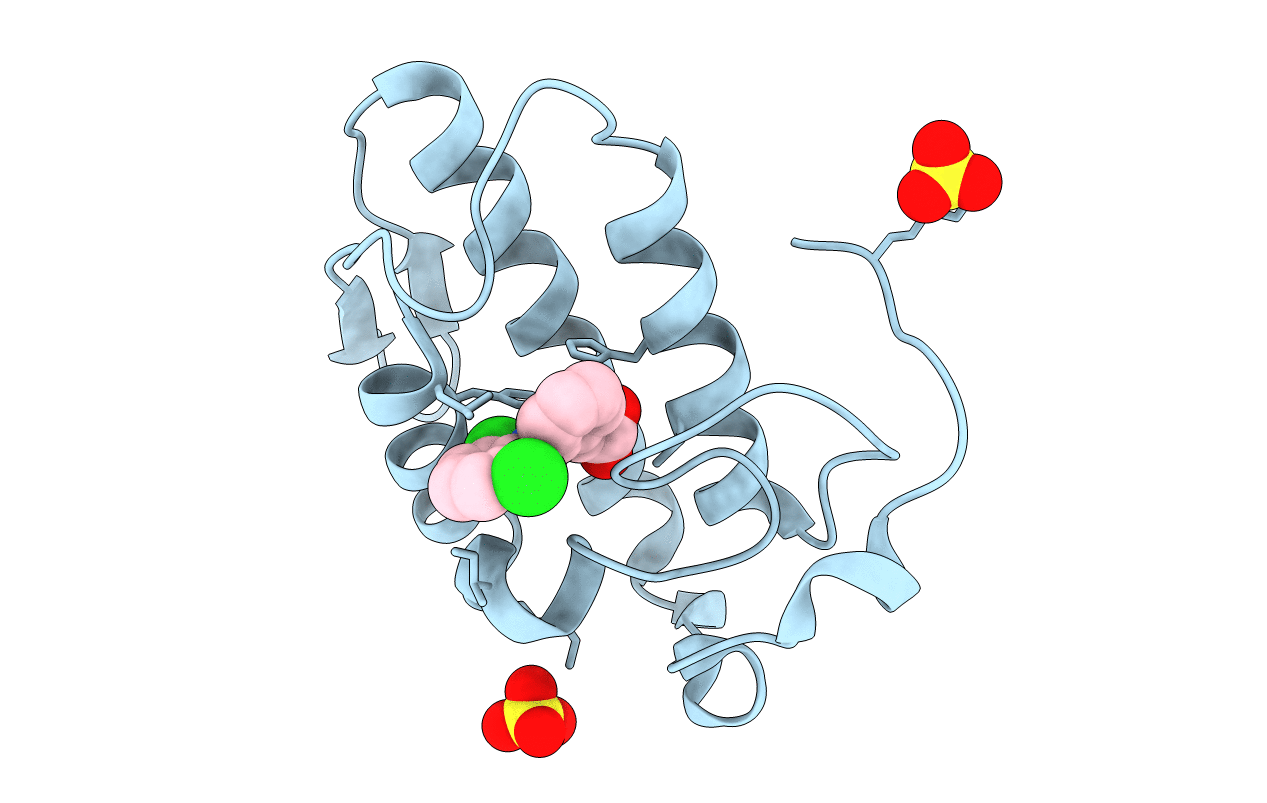
Specific binding of non-steroidal anti-inflammatory drugs (NSAIDs) to phospholipase A2: Crystal structure of the complex formed between phospholipase A2 and diclofenac at 2.7 A resolution:
Biological Source:
Source Organism(s):
Daboia russellii pulchella (Taxon ID: 97228)
Method Details:
Experimental Method:
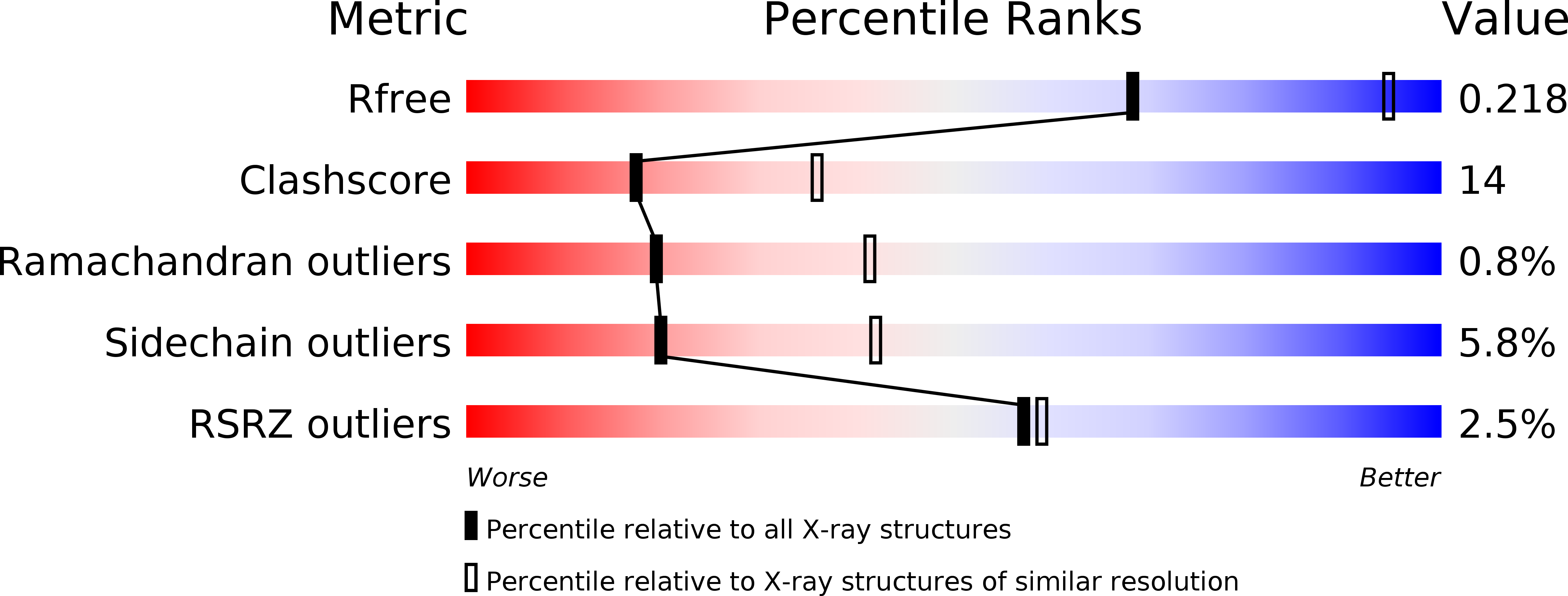
Resolution:
2.71 Å
R-Value Free:
0.21
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 43