Deposition Date
2005-08-31
Release Date
2006-02-07
Last Version Date
2024-02-14
Entry Detail
PDB ID:
2AWE
Keywords:
Title:
Base-Tetrad Swapping Results in Dimerization of RNA Quadruplexes: Implications for Formation of I-Motif RNA Octaplex
Method Details:
Experimental Method:
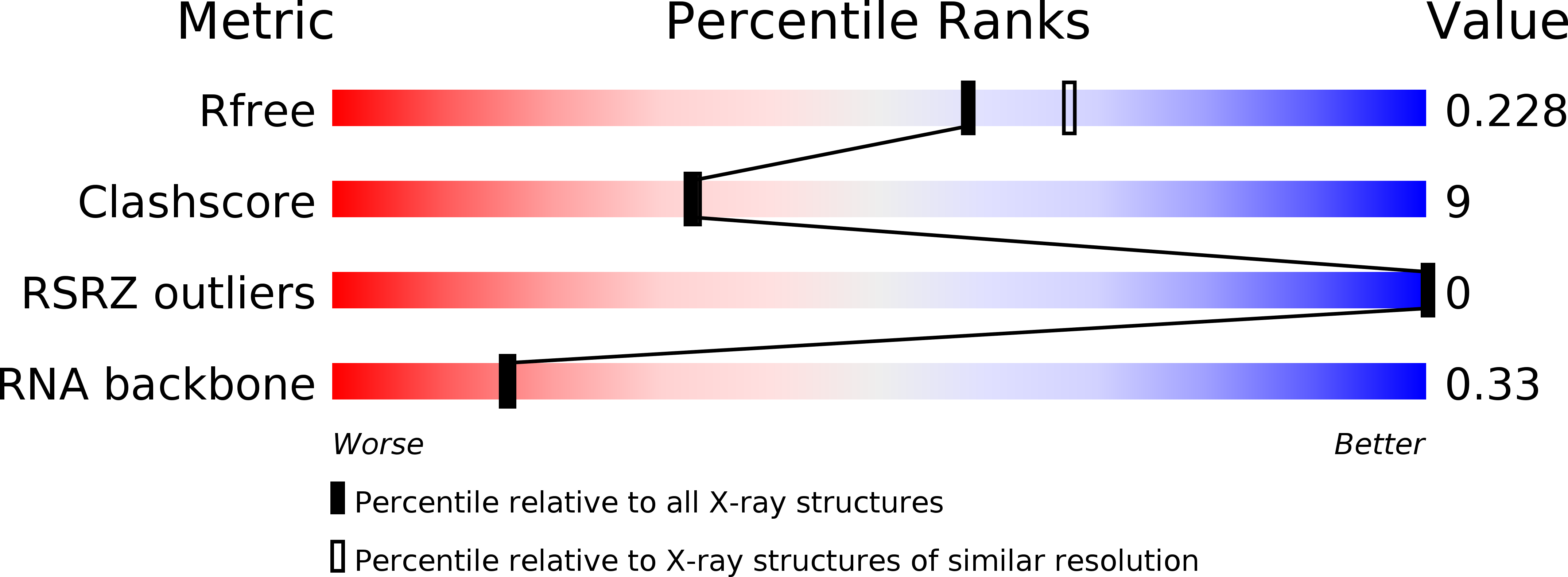
Resolution:
2.10 Å
R-Value Free:
0.22
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
I 41 2 2