Deposition Date
2005-08-30
Release Date
2006-01-24
Last Version Date
2023-08-23
Entry Detail
PDB ID:
2AVS
Keywords:
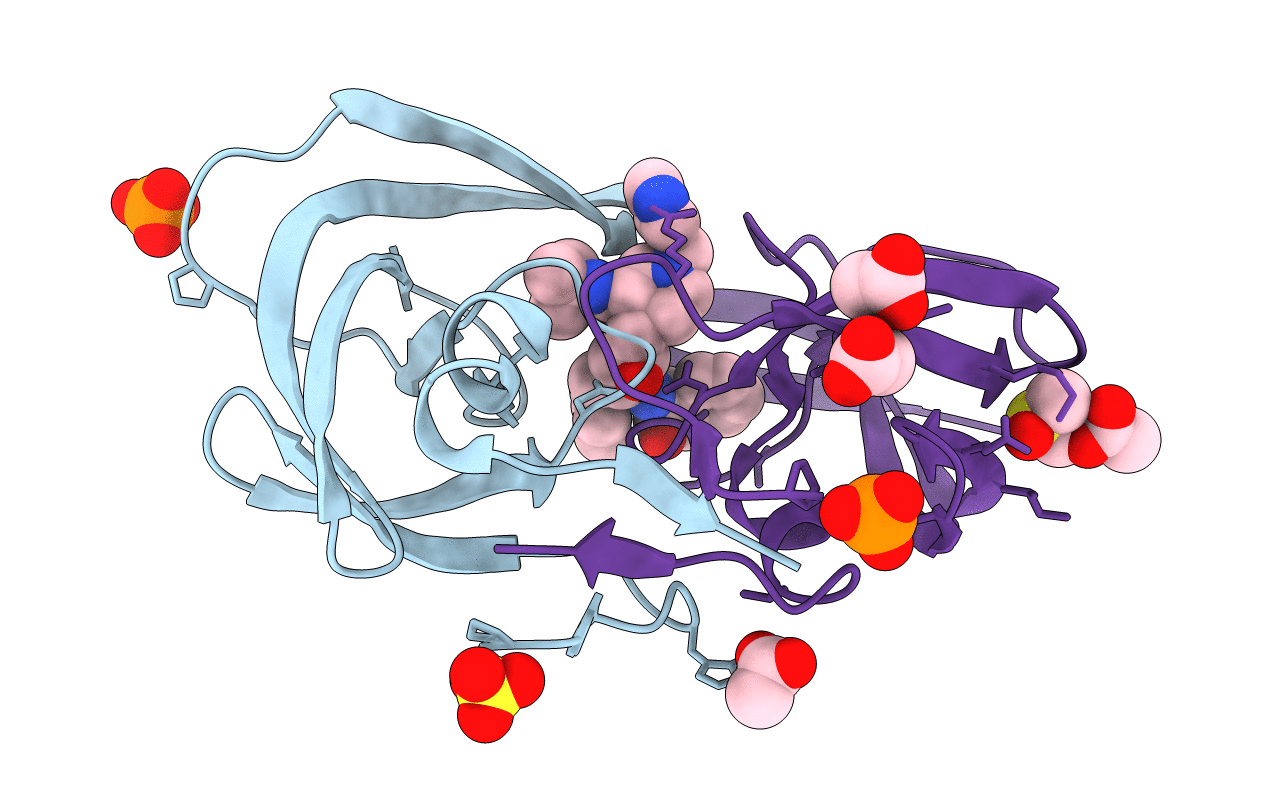
Title:
kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S
Biological Source:
Source Organism(s):
Human immunodeficiency virus 1 (Taxon ID: 11676)
Expression System(s):
Method Details:
Experimental Method:
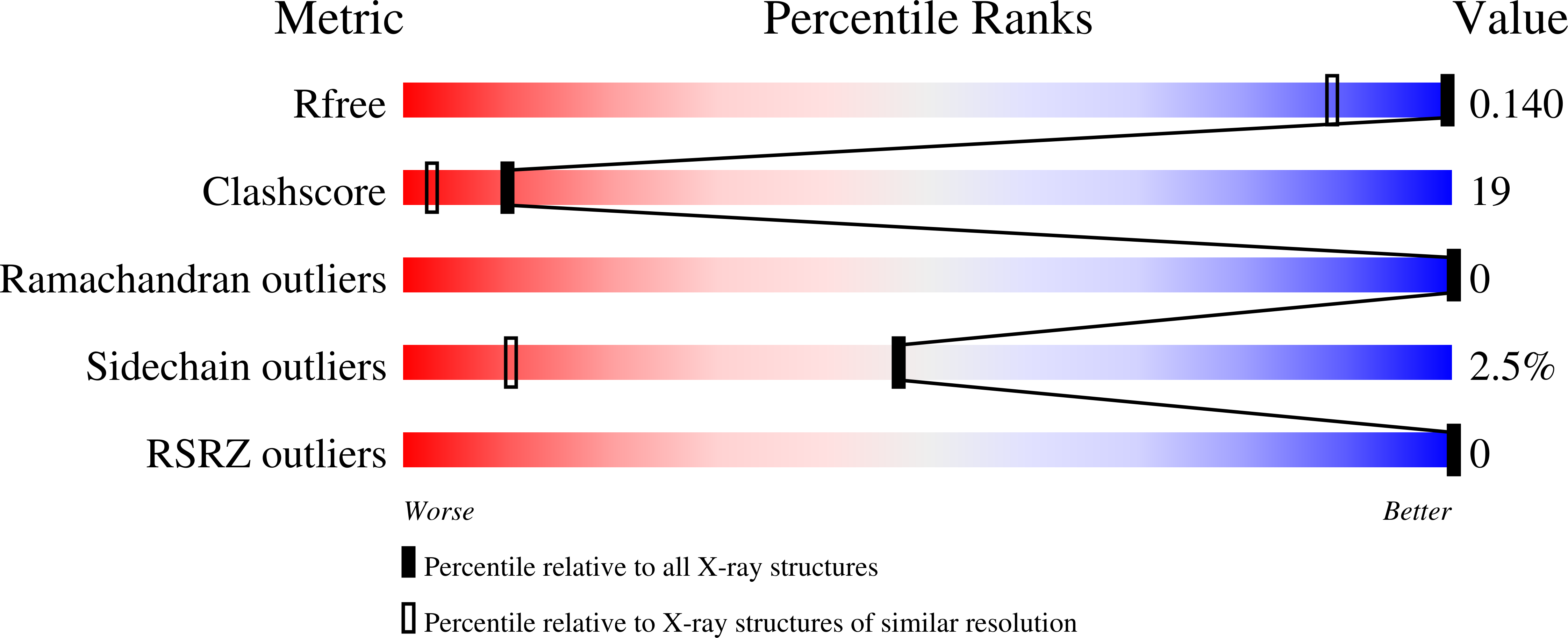
Resolution:
1.10 Å
R-Value Free:
0.14
R-Value Work:
0.10
R-Value Observed:
0.10
Space Group:
P 21 21 21