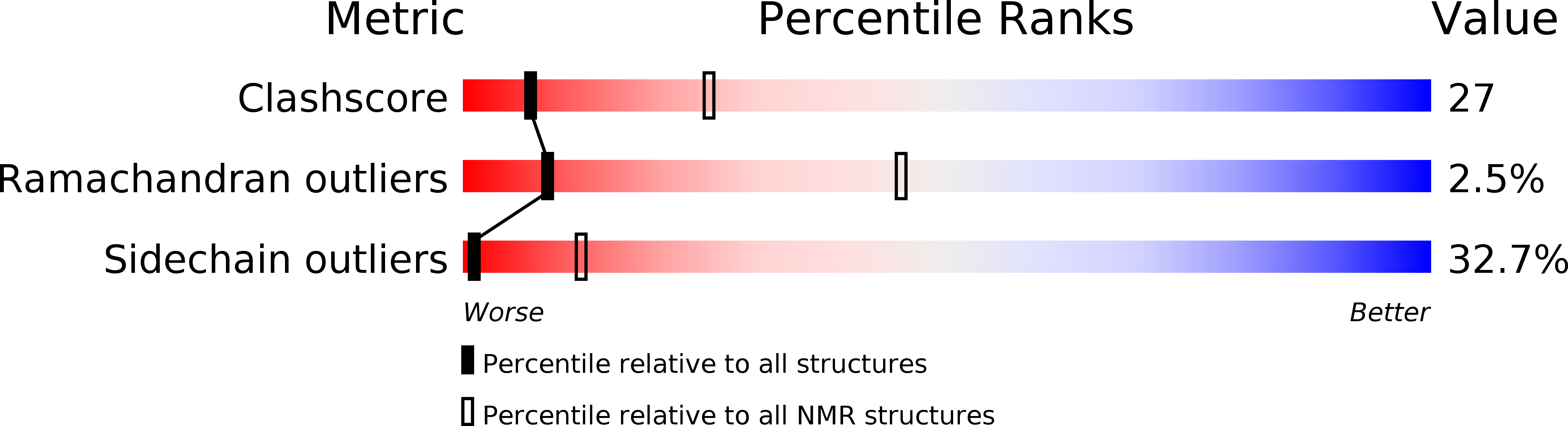Deposition Date
2005-08-01
Release Date
2006-03-14
Last Version Date
2024-10-30
Entry Detail
PDB ID:
2AIZ
Keywords:
Title:
Solution structure of peptidoglycan associated lipoprotein from Haemophilus influenza bound to UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine
Biological Source:
Source Organism(s):
Haemophilus influenzae (Taxon ID: 727)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the least restraint violations