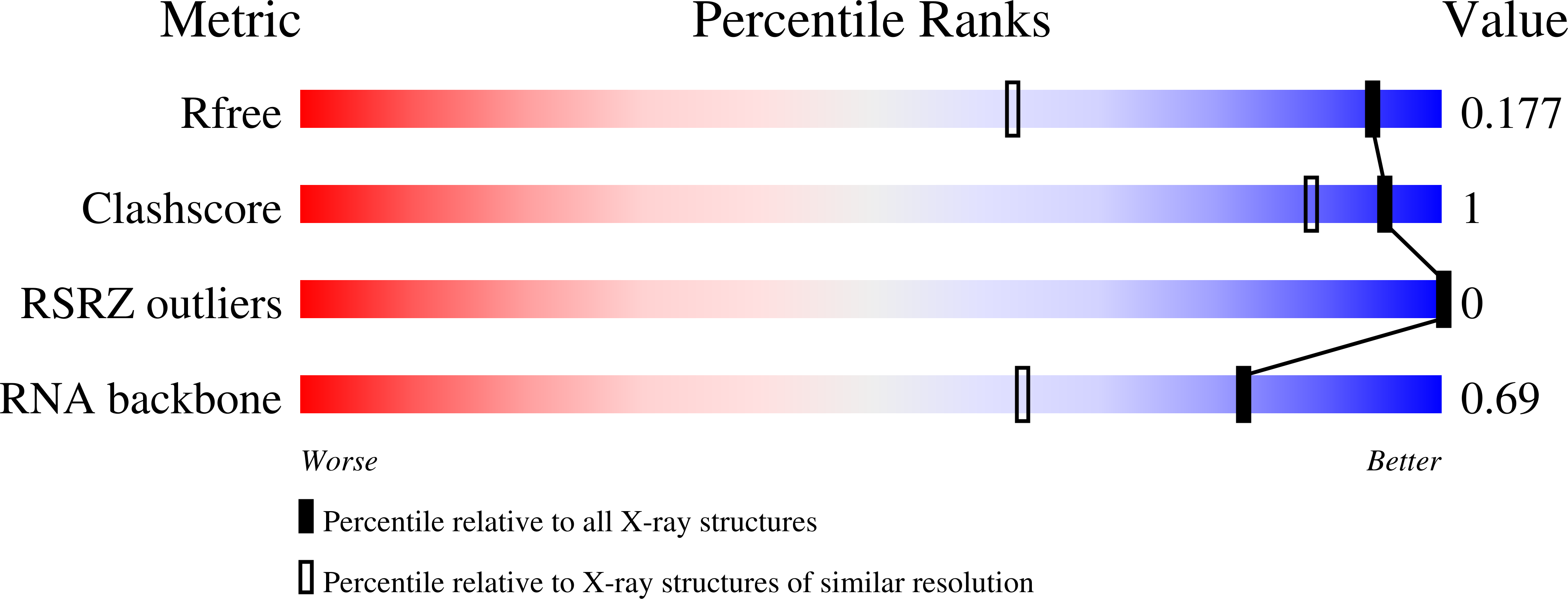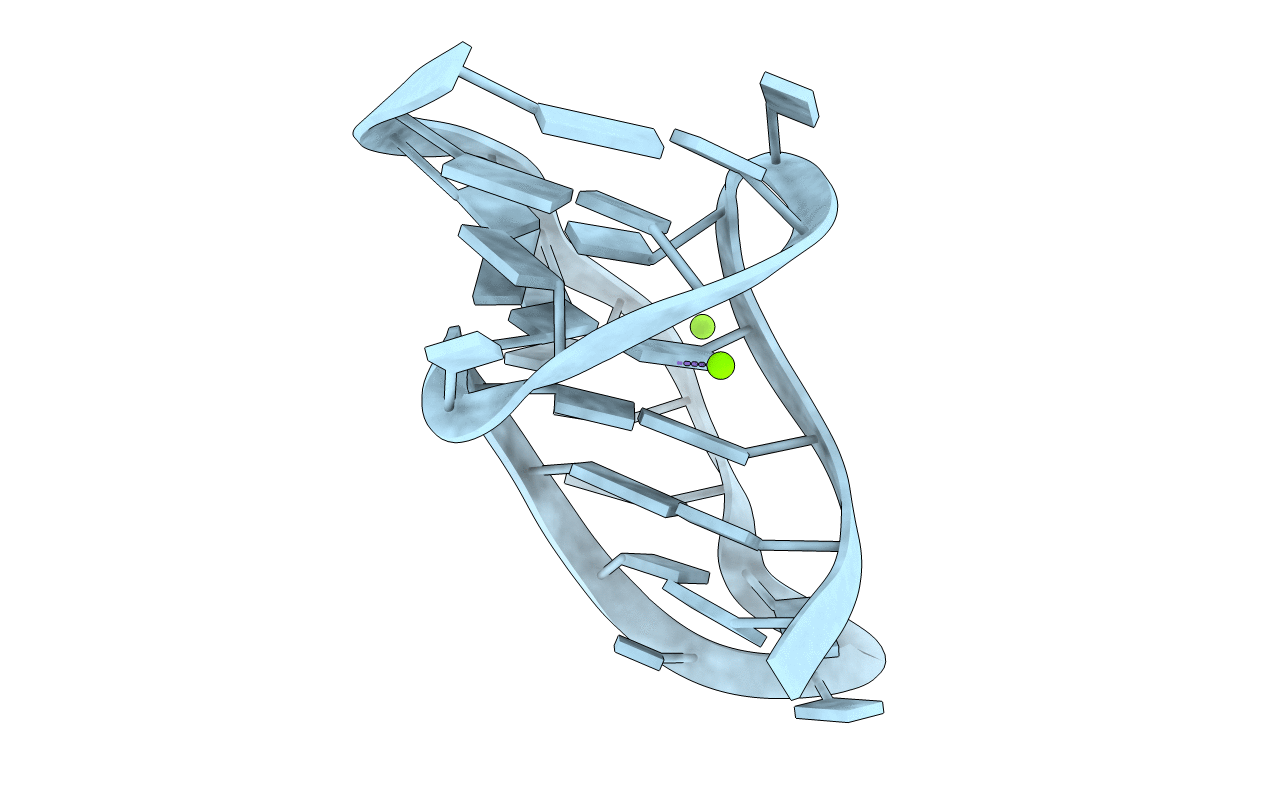
Deposition Date
2005-06-27
Release Date
2005-09-27
Last Version Date
2023-08-23
Entry Detail
PDB ID:
2A43
Keywords:
Title:
Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif
Method Details: