Deposition Date
1995-07-07
Release Date
1995-10-15
Last Version Date
2024-05-22
Entry Detail
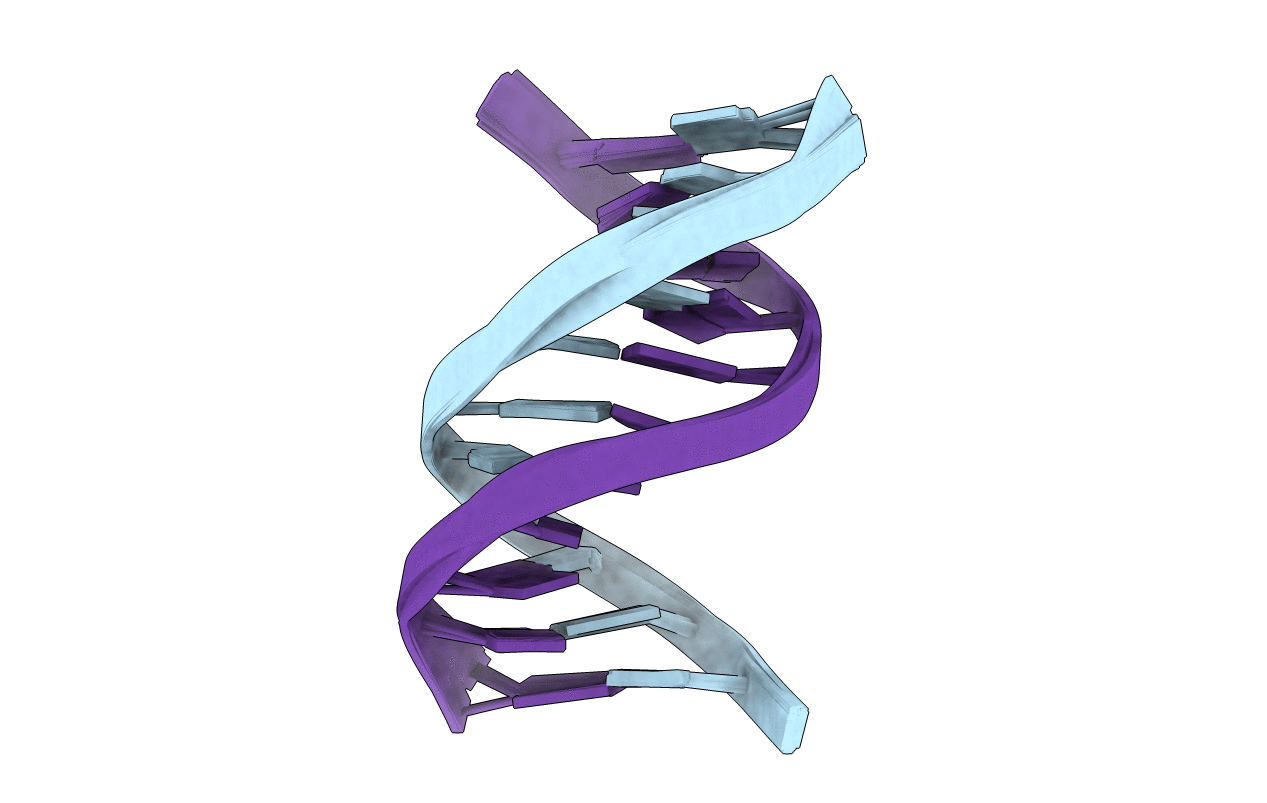
PDB ID:
214D
Keywords:
Title:
THE SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING A SINGLE 2'-O-METHYL-BETA-D-ARAT
Method Details:
Experimental Method:
Conformers Calculated:
40
Conformers Submitted:
40
Selection Criteria:
all calculated structures submitted


