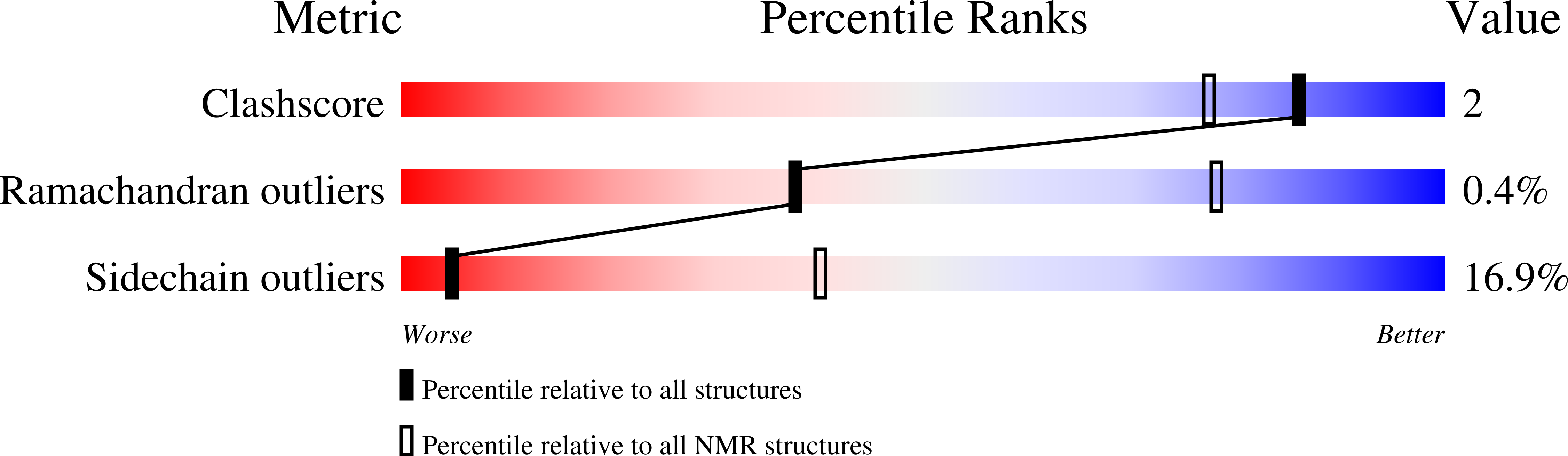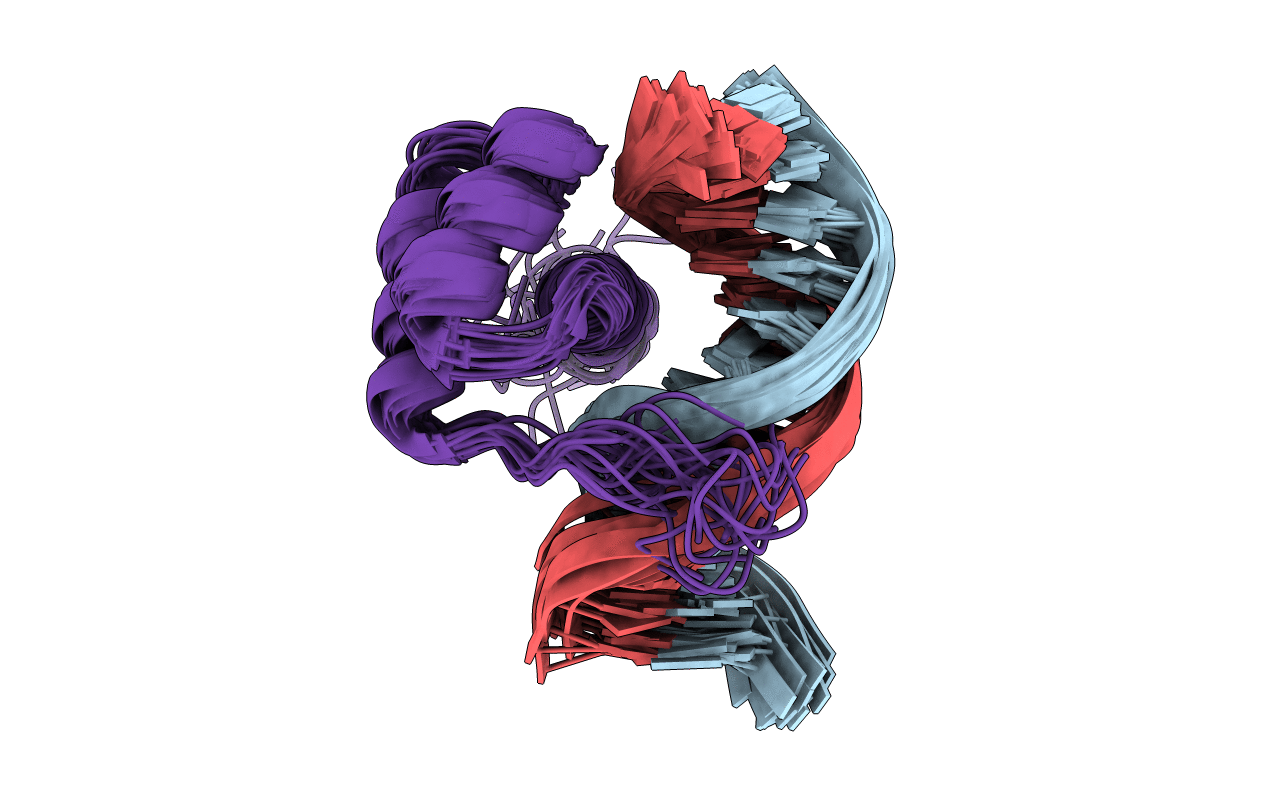
Deposition Date
2005-05-18
Release Date
2006-02-14
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1ZQ3
Keywords:
Title:
NMR Solution Structure of the Bicoid Homeodomain Bound to the Consensus DNA Binding Site TAATCC
Biological Source:
Source Organism:
Drosophila melanogaster (Taxon ID: 7227)
Host Organism:
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy