Deposition Date
2005-05-16
Release Date
2005-09-20
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1ZP7
Keywords:
Title:
The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence specific cleavage.
Biological Source:
Source Organism(s):
Bacillus subtilis (Taxon ID: 1423)
Method Details:
Experimental Method:
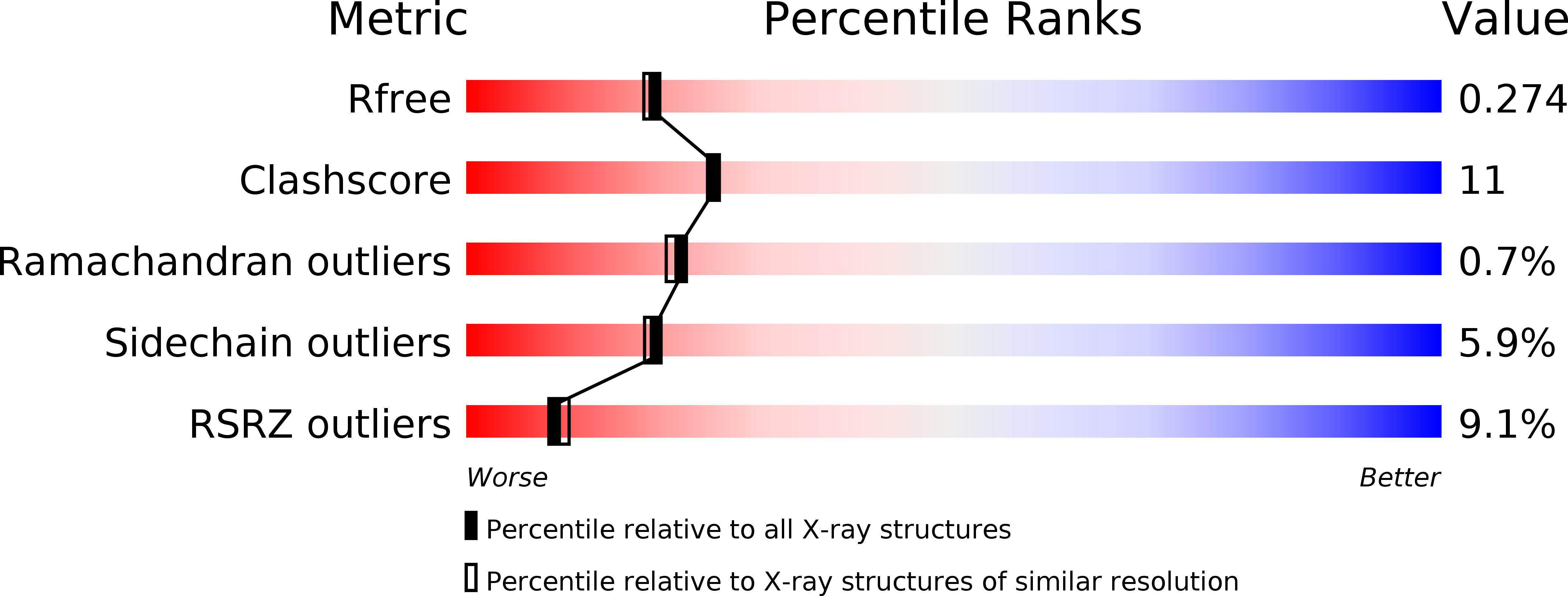
Resolution:
2.25 Å
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 65