Deposition Date
2005-04-22
Release Date
2005-08-02
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1ZH7
Keywords:
Title:
Structural and Biochemical Basis for Selective Repression of the Orphan Nuclear Receptor LRH-1 by SHP
Biological Source:
Source Organism(s):
Mus musculus (Taxon ID: 10090)
Rattus norvegicus (Taxon ID: 10116)
Rattus norvegicus (Taxon ID: 10116)
Method Details:
Experimental Method:
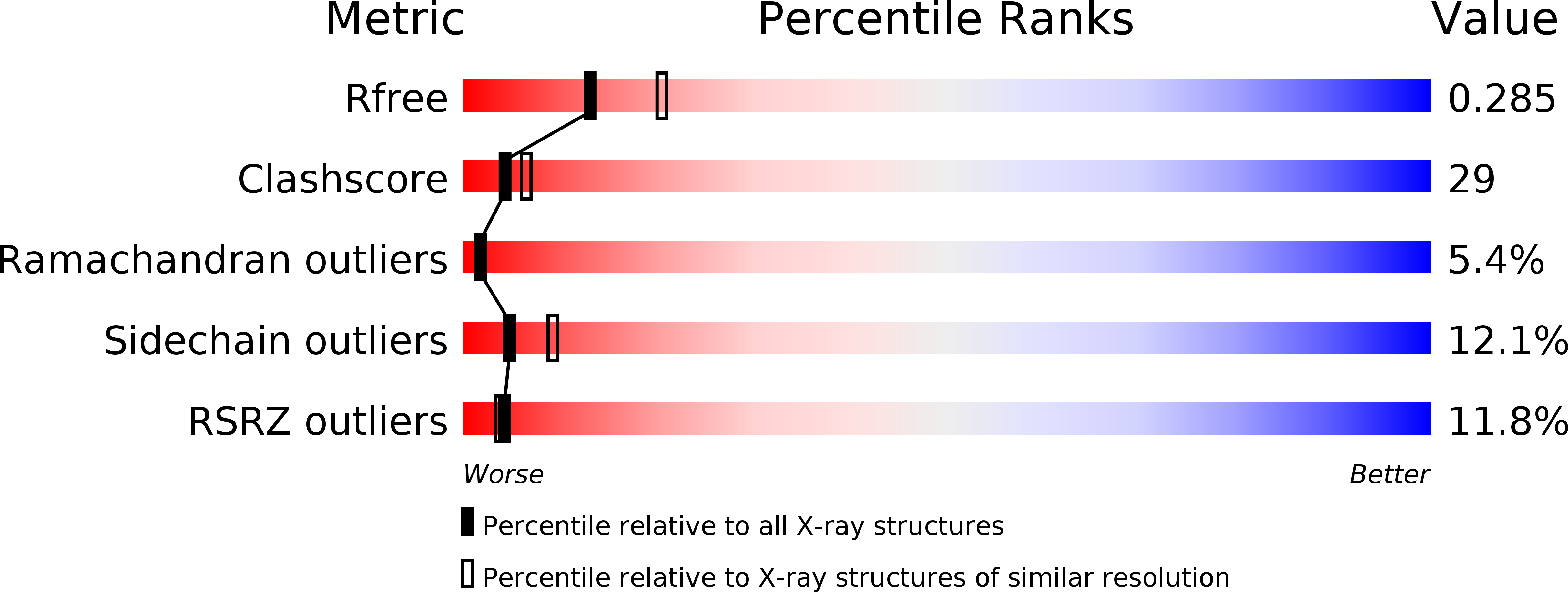
Resolution:
2.50 Å
R-Value Free:
0.28
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
C 1 2 1