Deposition Date
2005-04-22
Release Date
2005-10-18
Last Version Date
2024-11-06
Entry Detail
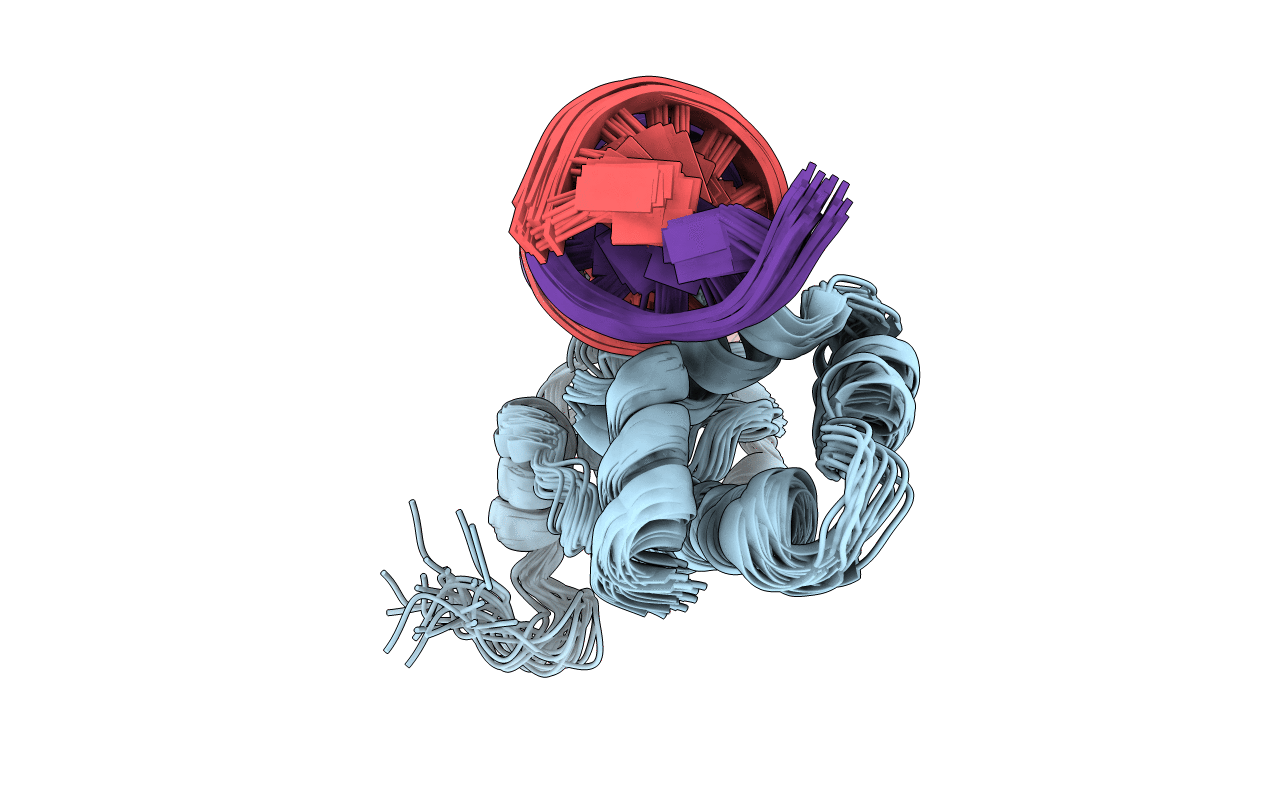
PDB ID:
1ZGW
Keywords:
Title:
NMR structure of E. Coli Ada protein in complex with DNA
Biological Source:
Source Organism:
Escherichia coli (Taxon ID: 562)
Host Organism:
Method Details:
Experimental Method:
Conformers Calculated:
40
Conformers Submitted:
20
Selection Criteria:
The submitted conformer models have no restraint violations and lowest energies


