Deposition Date
2005-02-28
Release Date
2005-10-18
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1YZD
Keywords:
Title:
Crystal structure of an RNA duplex containing a site specific 2'-amine substitution at a C-G Watson-Crick base pair
Method Details:
Experimental Method:
Resolution:
2.35 Å
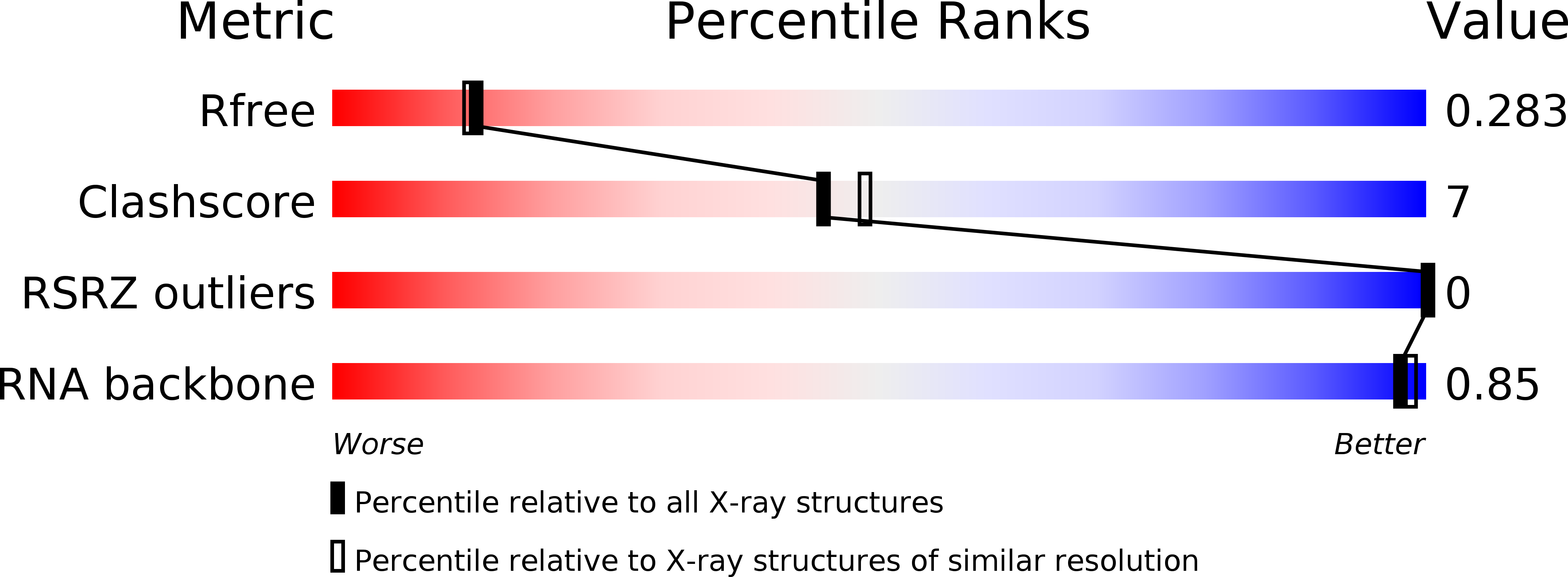
R-Value Free:
0.29
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 31 2 1