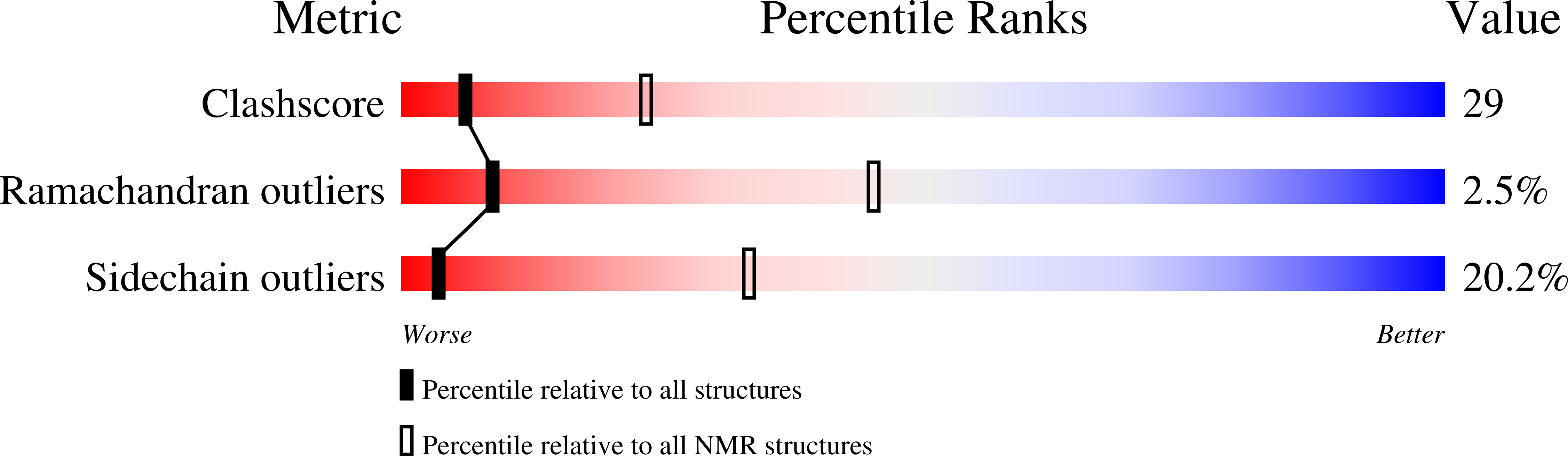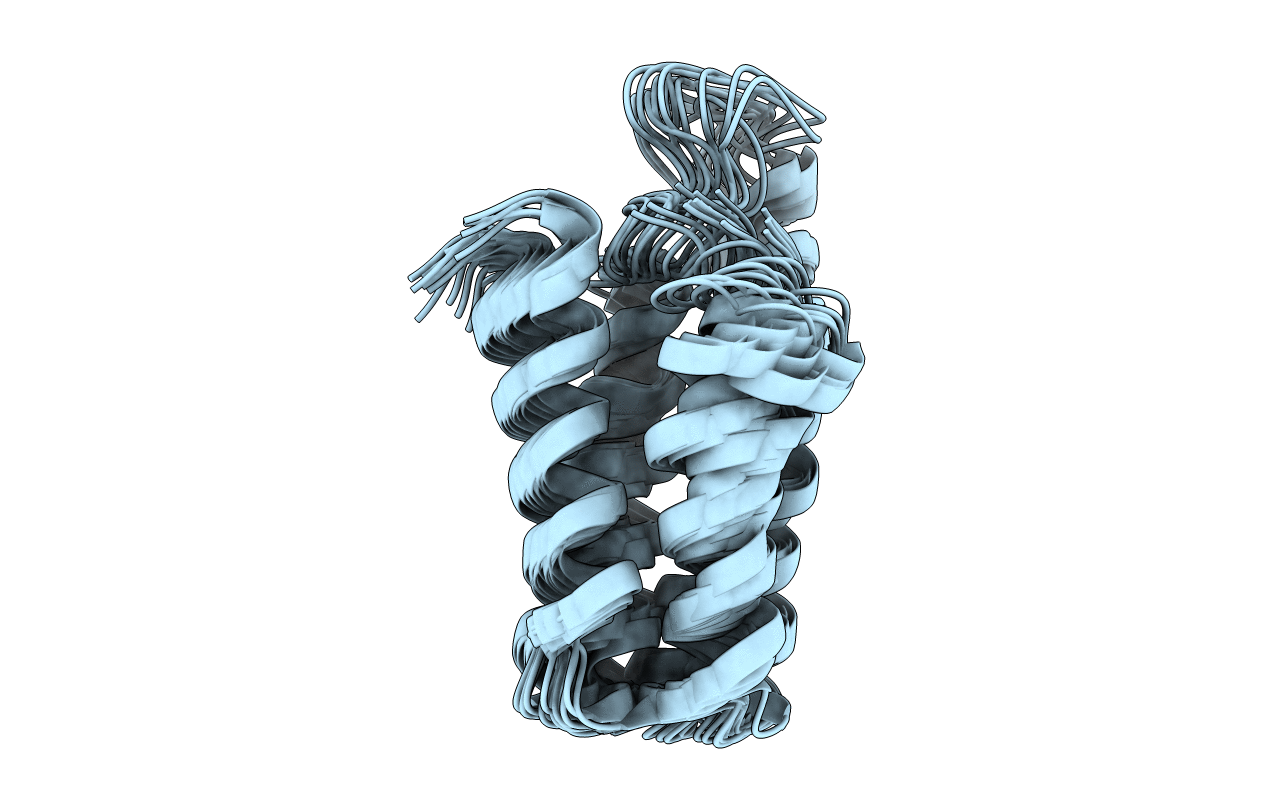
Deposition Date
2005-02-25
Release Date
2005-08-25
Last Version Date
2024-05-29
Entry Detail
PDB ID:
1YYX
Keywords:
Title:
The solution structure of a redesigned apocytochrome B562 (Rd-apocyt b562) at 2.8M urea
Method Details: