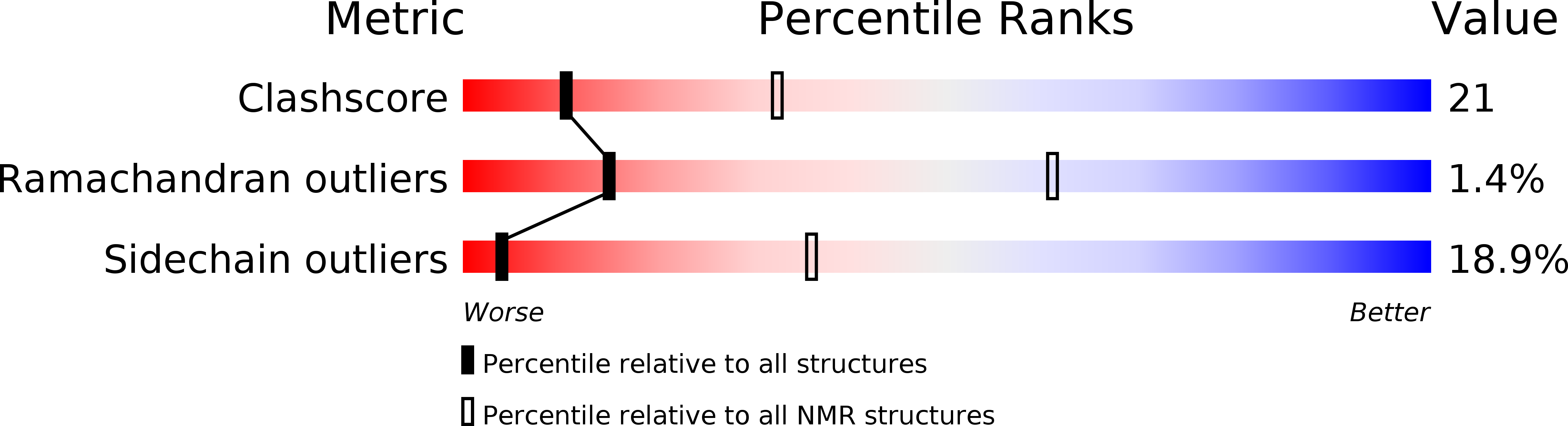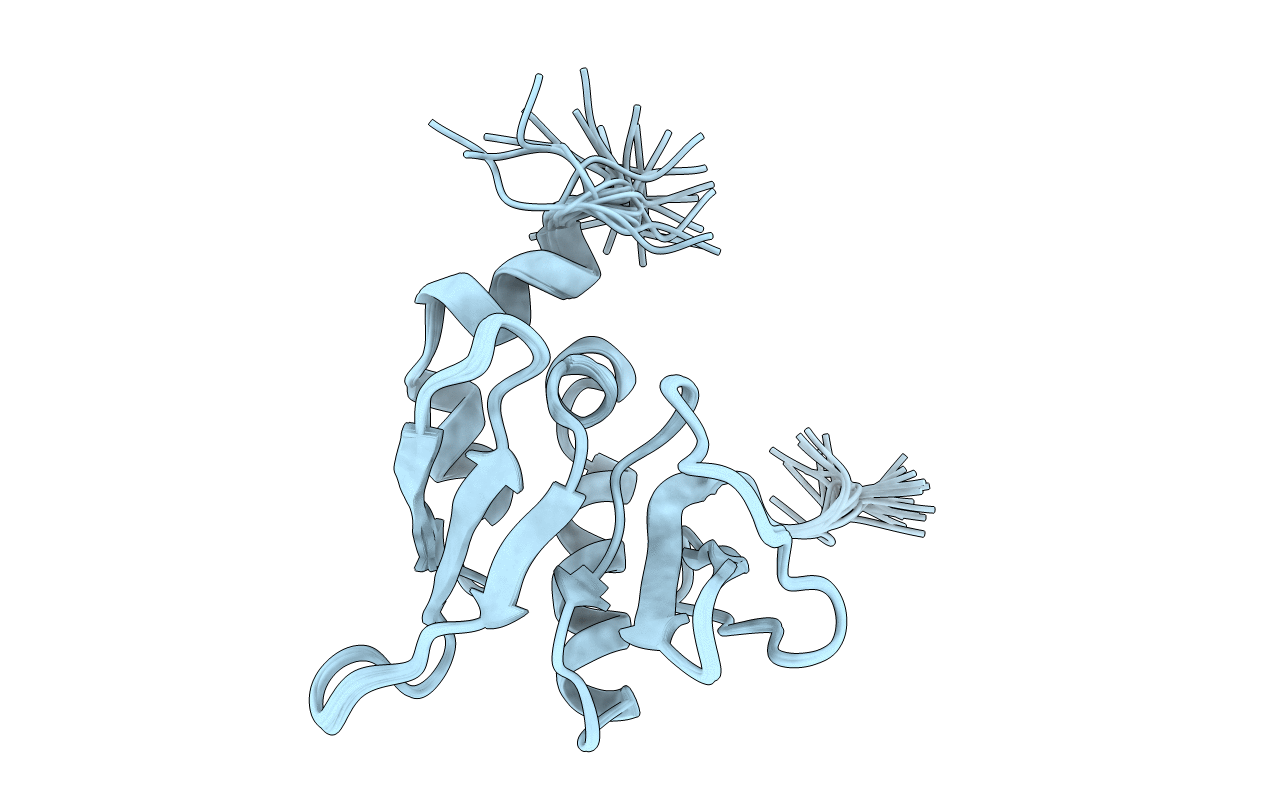
Deposition Date
2005-02-02
Release Date
2006-02-07
Last Version Date
2024-05-01
Entry Detail
PDB ID:
1YR1
Keywords:
Title:
Structure of the major extracytoplasmic domain of the trans isomer of the bacterial cell division protein divib from geobacillus stearothermophilus
Biological Source:
Source Organism(s):
Geobacillus stearothermophilus (Taxon ID: 1422)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
60
Conformers Submitted:
25
Selection Criteria:
lowest energy