Deposition Date
2005-01-26
Release Date
2005-03-01
Last Version Date
2023-08-23
Entry Detail
PDB ID:
1YNY
Keywords:
Title:
Molecular Structure of D-Hydantoinase from a Bacillus sp. AR9: Evidence for mercury inhibition
Biological Source:
Source Organism(s):
Bacillus sp. (Taxon ID: 301298)
Expression System(s):
Method Details:
Experimental Method:
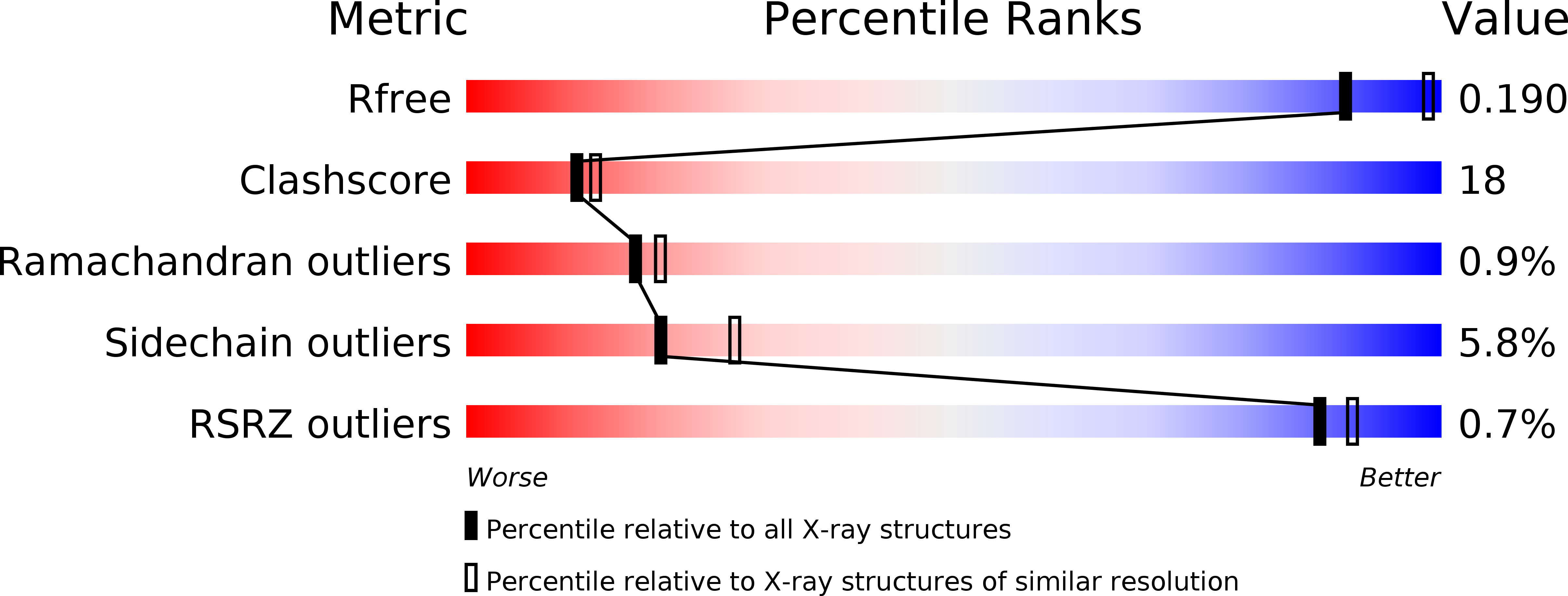
Resolution:
2.30 Å
R-Value Free:
0.24
R-Value Work:
0.19
Space Group:
P 64