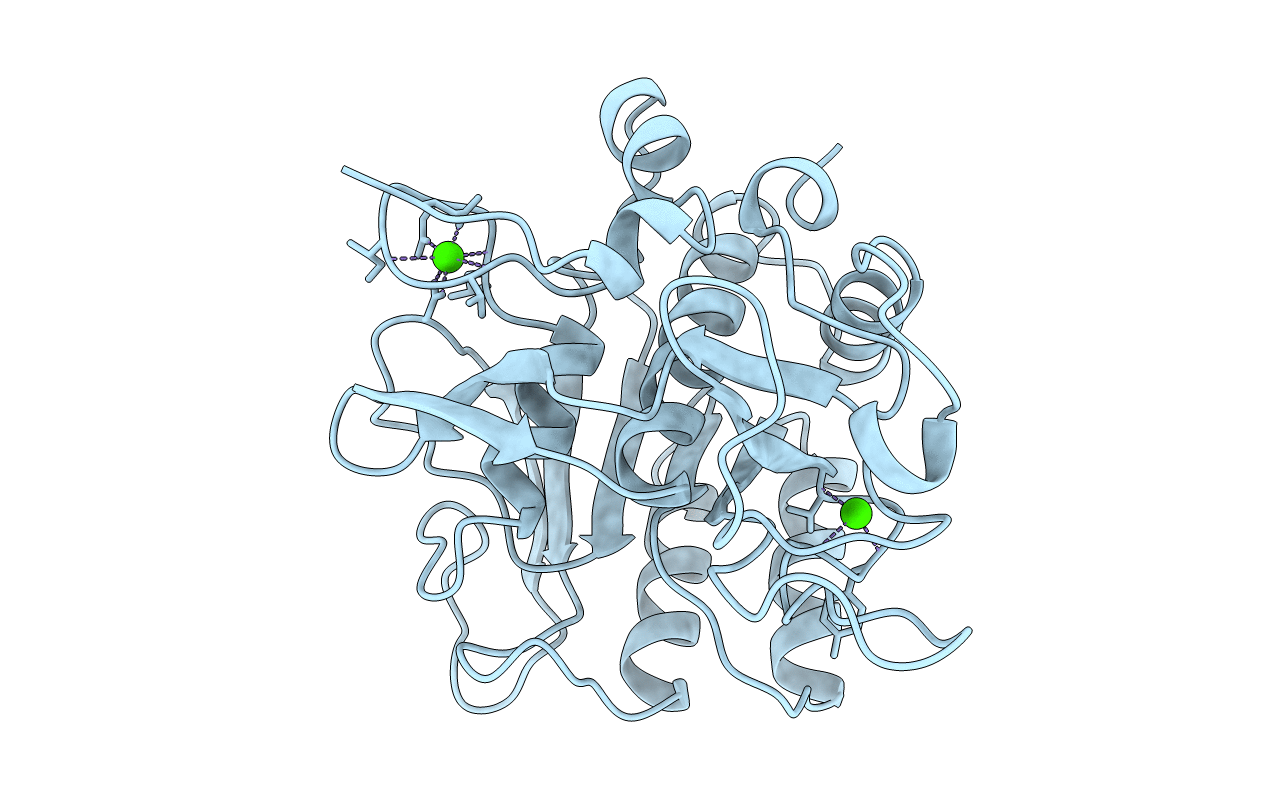
Deposition Date
1996-01-16
Release Date
1996-07-11
Last Version Date
2024-10-23
Entry Detail
PDB ID:
1YJB
Keywords:
Title:
SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 35% DIMETHYLFORMAMIDE
Biological Source:
Source Organism(s):
Bacillus amyloliquefaciens (Taxon ID: 1390)
Expression System(s):
Method Details:
Experimental Method:
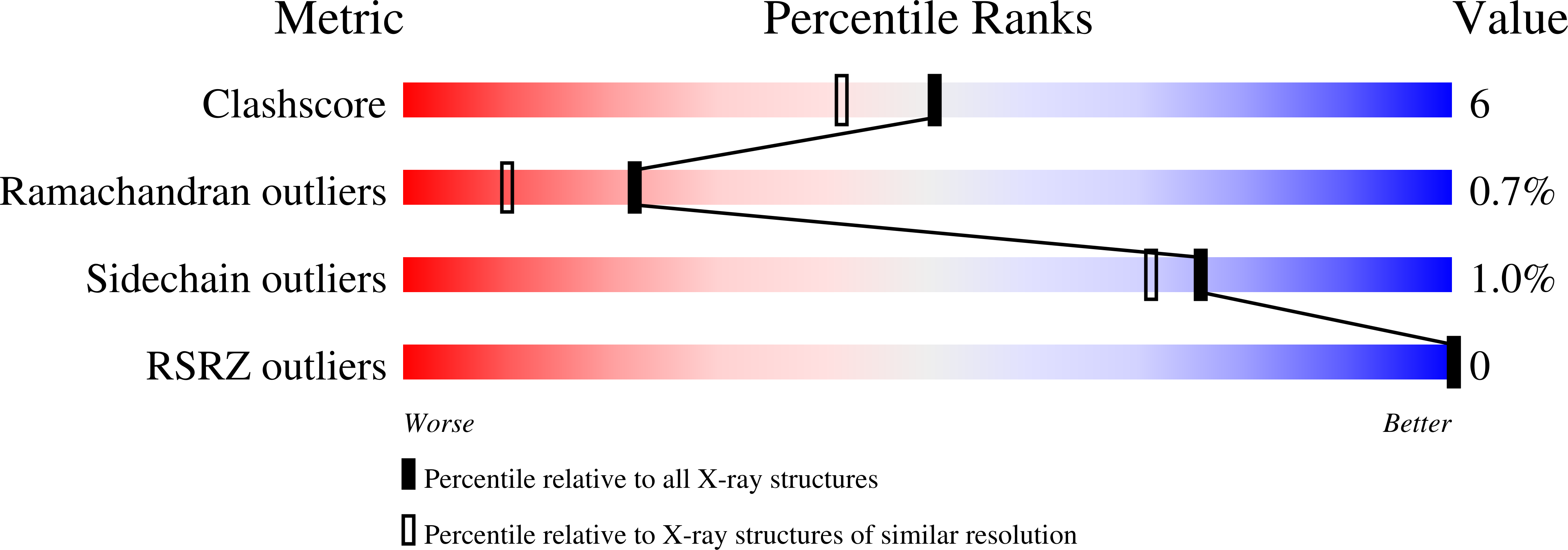
Resolution:
1.80 Å
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 1 21 1