Deposition Date
2005-01-12
Release Date
2005-04-26
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1YIJ
Keywords:
Title:
Crystal Structure Of Telithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui
Biological Source:
Source Organism:
Haloarcula marismortui (Taxon ID: 2238)
Method Details:
Experimental Method:
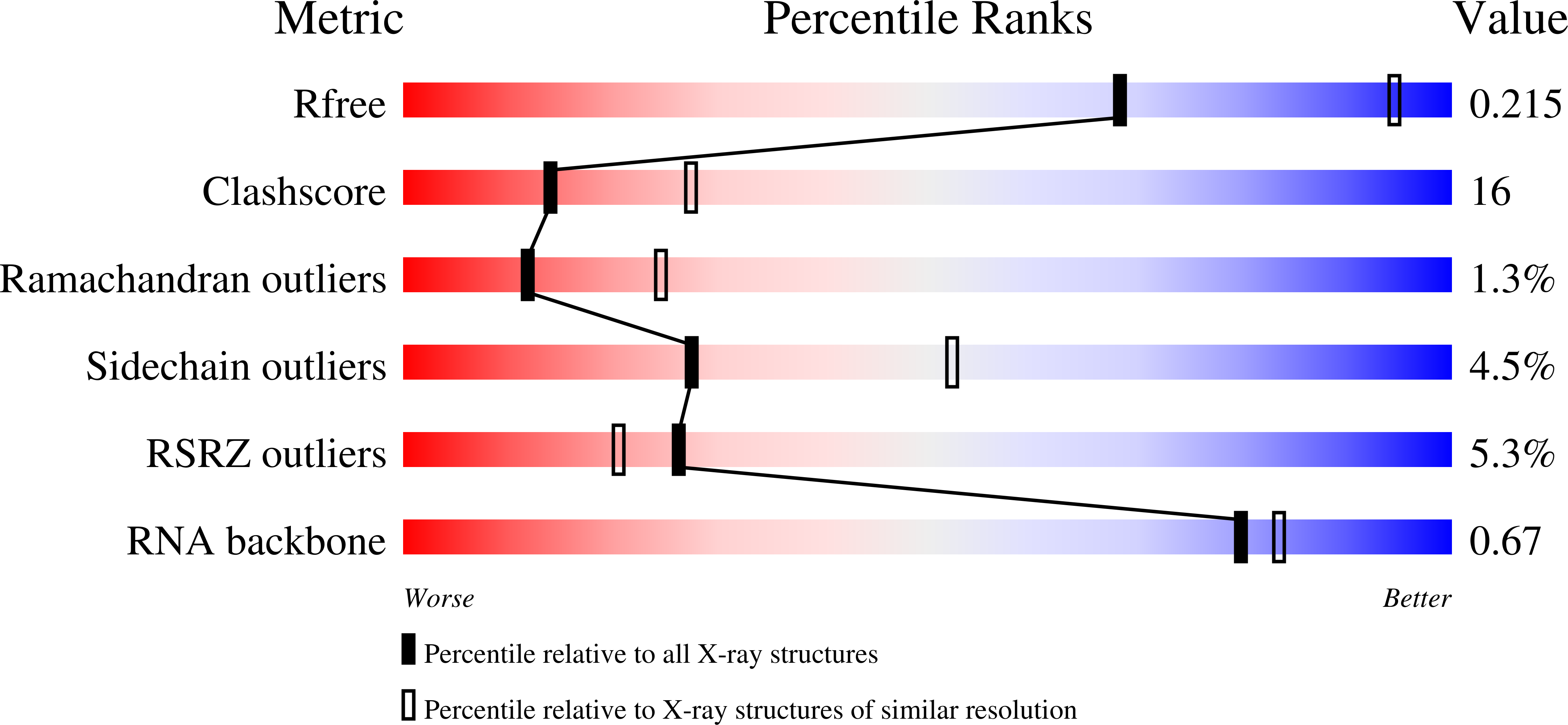
Resolution:
2.60 Å
R-Value Free:
0.22
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
C 2 2 21