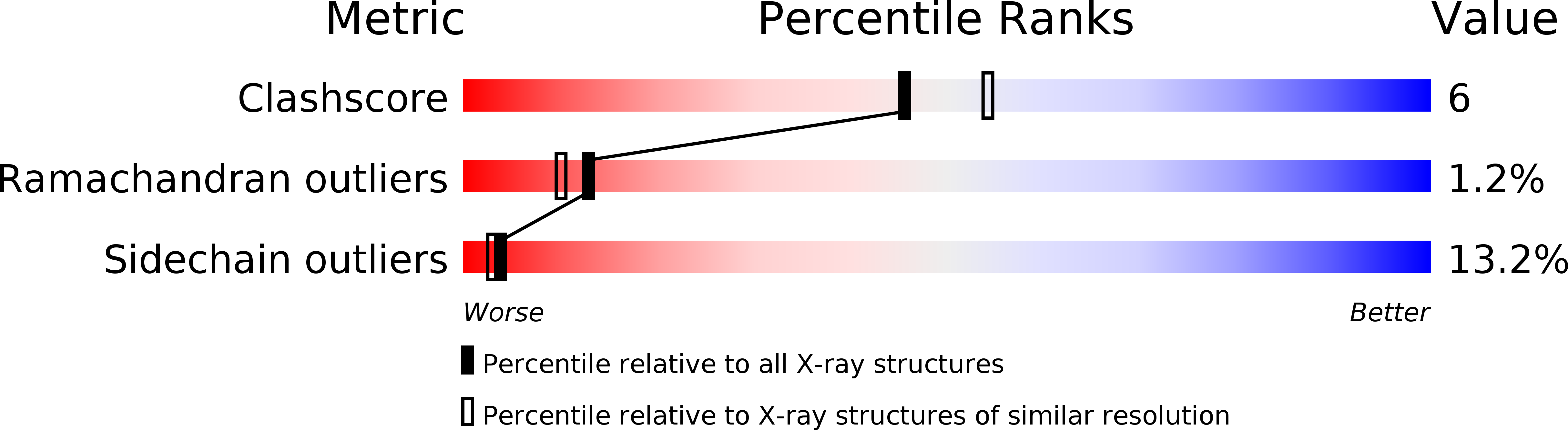
Deposition Date
1994-04-14
Release Date
1994-06-22
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1YHB
Keywords:
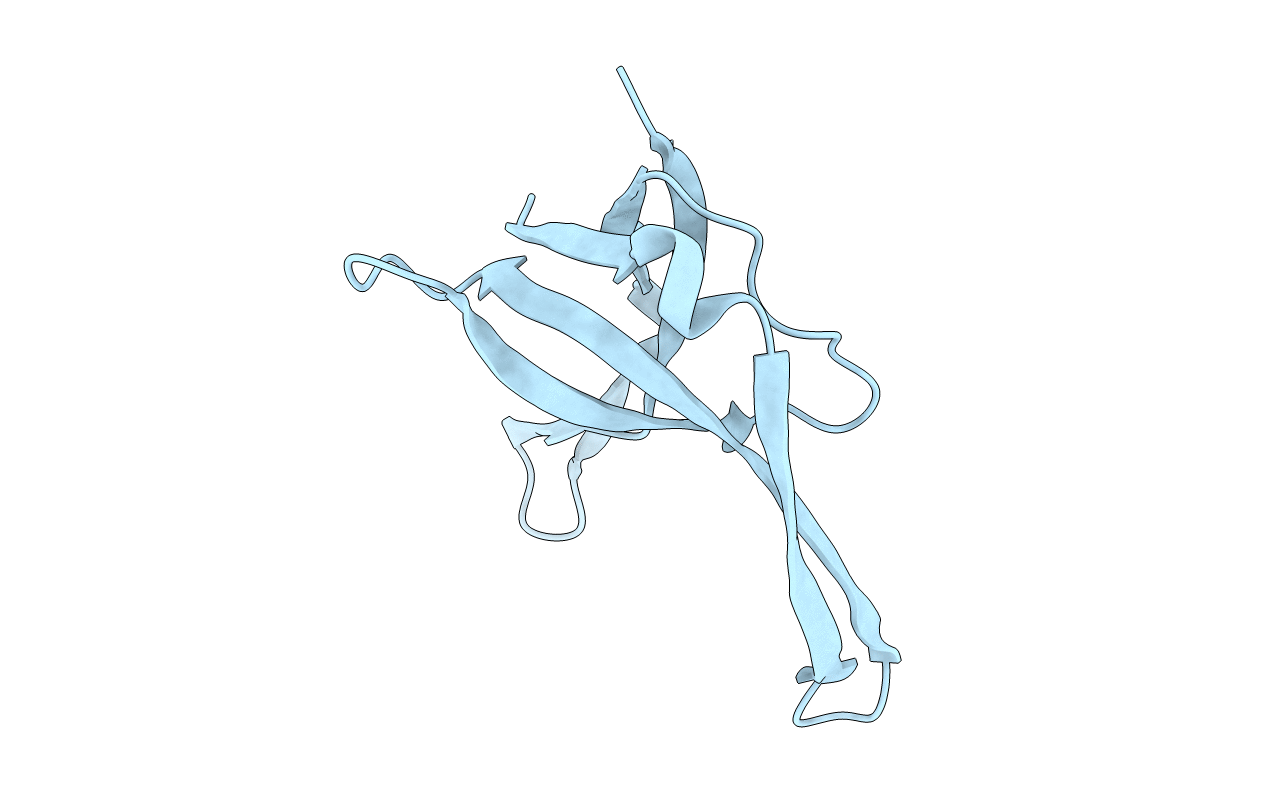
Title:
CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX
Biological Source:
Source Organism(s):
Enterobacteria phage f1 (Taxon ID: 10863)
Method Details: