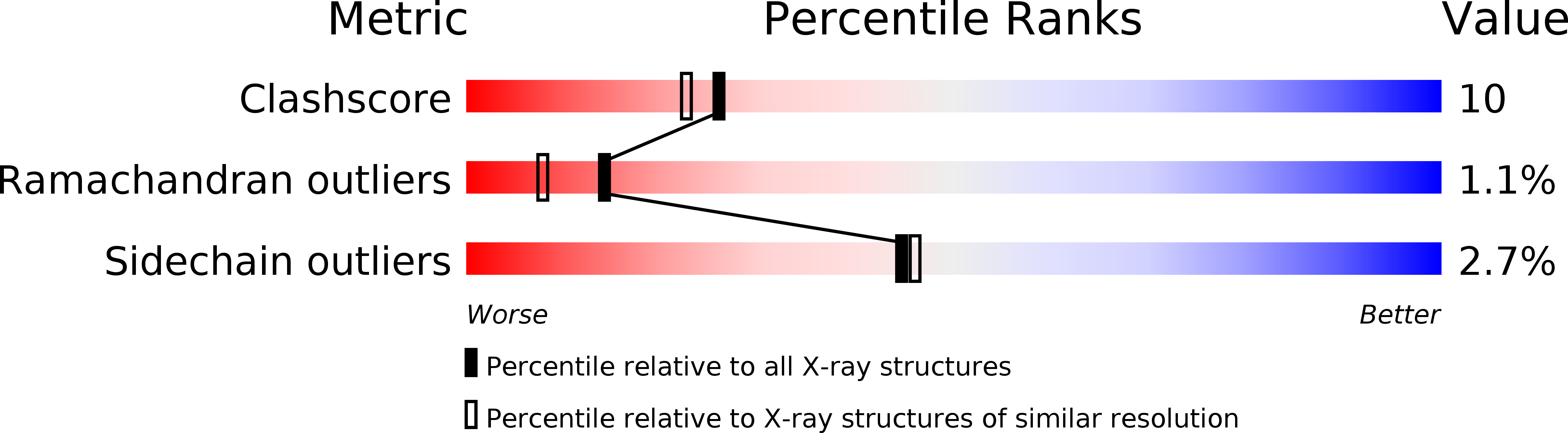
Deposition Date
1994-08-09
Release Date
1995-08-08
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1XYN
Keywords:
Title:
STRUCTURAL COMPARISON OF TWO MAJOR ENDO-1,4-BETA-XYLANASES FROM TRICHODREMA REESEI
Biological Source:
Source Organism(s):
Hypocrea jecorina (Taxon ID: 51453)
Method Details:
Experimental Method:
Resolution:
2.00 Å
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
C 1 2 1