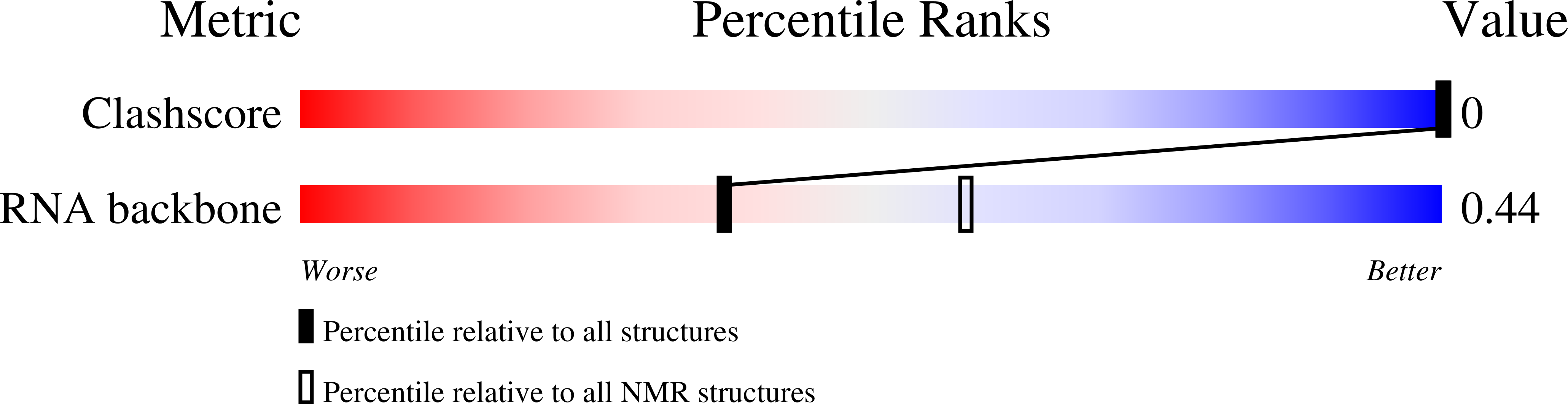
Deposition Date
2004-10-26
Release Date
2004-11-09
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1XV0
Keywords:
Title:
Solution NMR structure of RNA internal loop with three consecutive sheared GA pairs in 5'GGUGGAGGCU/3'PCCGAAGCCG
Method Details:
Experimental Method:
Conformers Calculated:
40
Conformers Submitted:
33
Selection Criteria:
structures with the least restraint violations