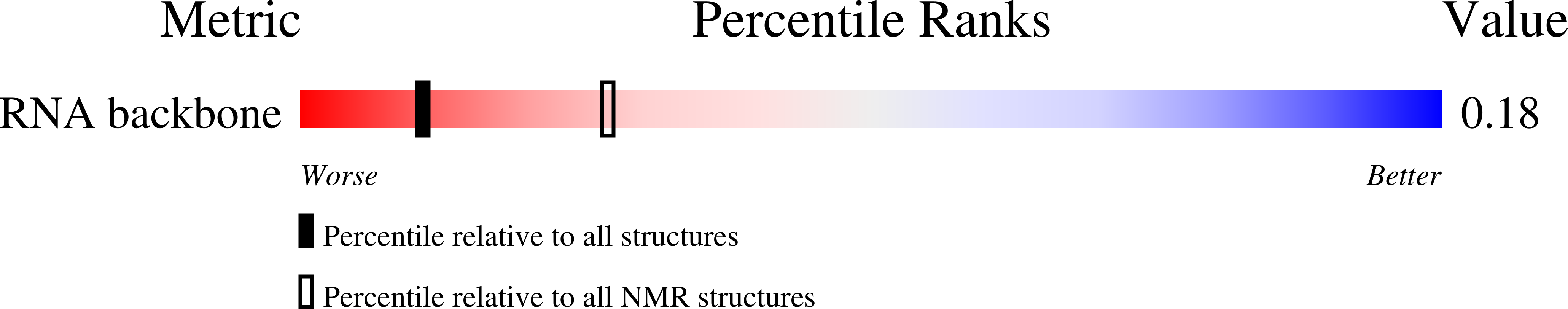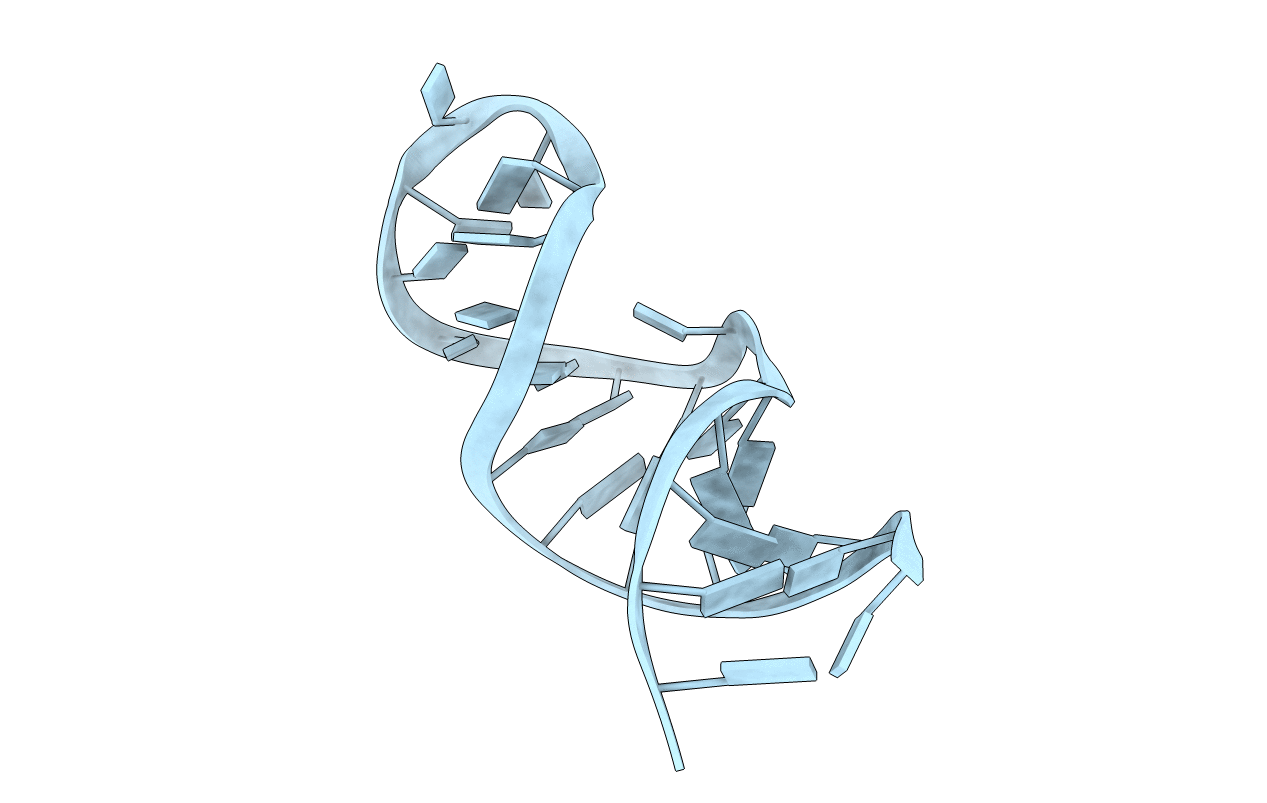
Deposition Date
2004-10-19
Release Date
2004-12-28
Last Version Date
2024-05-29
Entry Detail
PDB ID:
1XSH
Keywords:
Title:
Solution structure of E.coli RNase P RNA P4 stem oligoribonucleotide, U69C/C70U mutation
Method Details:
Experimental Method:
Conformers Submitted:
1