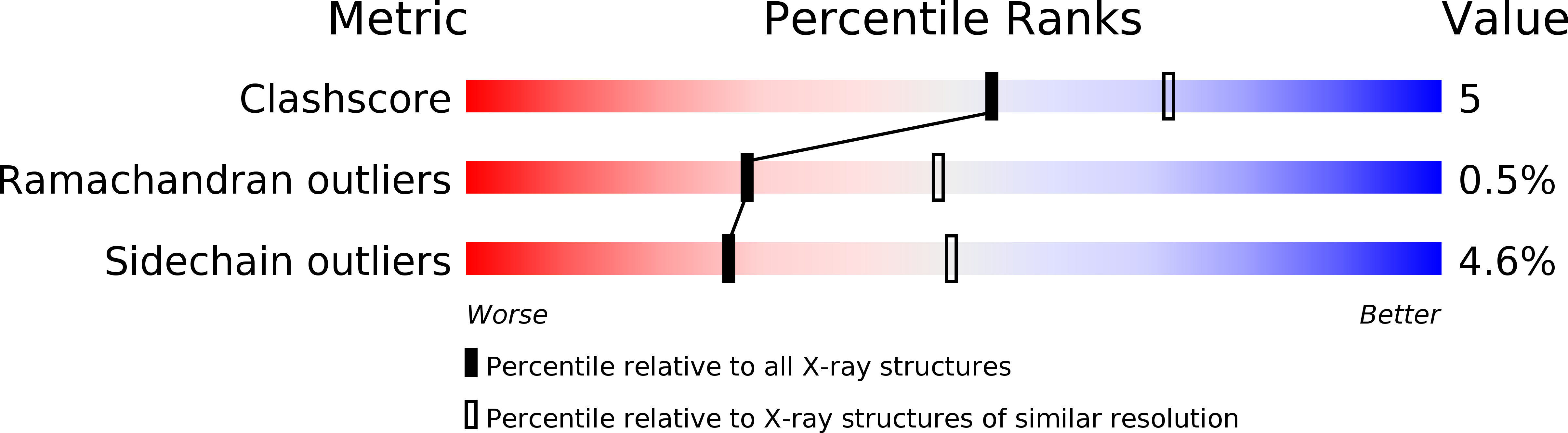Deposition Date
1991-10-09
Release Date
1993-07-15
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1XLE
Keywords:
Title:
MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT
Biological Source:
Source Organism(s):
Arthrobacter sp. (Taxon ID: 1669)
Method Details:
Experimental Method:
Resolution:
2.50 Å
R-Value Observed:
0.17
Space Group:
P 31 2 1